tomography
-
 KAIST Develops Virtual Staining Technology for 3D Histopathology
Moving beyond traditional methods of observing thinly sliced and stained cancer tissues, a collaborative international research team led by KAIST has successfully developed a groundbreaking technology. This innovation uses advanced optical techniques combined with an artificial intelligence-based deep learning algorithm to create realistic, virtually stained 3D images of cancer tissue without the need for serial sectioning nor staining. This breakthrough is anticipated to pave the way for next-generation non-invasive pathological diagnosis.
< Photo 1. (From left) Juyeon Park (Ph.D. Candidate, Department of Physics), Professor YongKeun Park (Department of Physics) (Top left) Professor Su-Jin Shin (Gangnam Severance Hospital), Professor Tae Hyun Hwang (Vanderbilt University School of Medicine) >
KAIST (President Kwang Hyung Lee) announced on the 26th that a research team led by Professor YongKeun Park of the Department of Physics, in collaboration with Professor Su-Jin Shin's team at Yonsei University Gangnam Severance Hospital, Professor Tae Hyun Hwang's team at Mayo Clinic, and Tomocube's AI research team, has developed an innovative technology capable of vividly displaying the 3D structure of cancer tissues without separate staining.
For over 200 years, conventional pathology has relied on observing cancer tissues under a microscope, a method that only shows specific cross-sections of the 3D cancer tissue. This has limited the ability to understand the three-dimensional connections and spatial arrangements between cells.
To overcome this, the research team utilized holotomography (HT), an advanced optical technology, to measure the 3D refractive index information of tissues. They then integrated an AI-based deep learning algorithm to successfully generate virtual H&E* images.* H&E (Hematoxylin & Eosin): The most widely used staining method for observing pathological tissues. Hematoxylin stains cell nuclei blue, and eosin stains cytoplasm pink.
The research team quantitatively demonstrated that the images generated by this technology are highly similar to actual stained tissue images. Furthermore, the technology exhibited consistent performance across various organs and tissues, proving its versatility and reliability as a next-generation pathological analysis tool.
< Figure 1. Comparison of conventional 3D tissue pathology procedure and the 3D virtual H&E staining technology proposed in this study. The traditional method requires preparing and staining dozens of tissue slides, while the proposed technology can reduce the number of slides by up to 10 times and quickly generate H&E images without the staining process. >
Moreover, by validating the feasibility of this technology through joint research with hospitals and research institutions in Korea and the United States, utilizing Tomocube's holotomography equipment, the team demonstrated its potential for full-scale adoption in real-world pathological research settings.
Professor YongKeun Park stated, "This research marks a major advancement by transitioning pathological analysis from conventional 2D methods to comprehensive 3D imaging. It will greatly enhance biomedical research and clinical diagnostics, particularly in understanding cancer tumor boundaries and the intricate spatial arrangements of cells within tumor microenvironments."
< Figure 2. Results of AI-based 3D virtual H&E staining and quantitative analysis of pathological tissue. The virtually stained images enabled 3D reconstruction of key pathological features such as cell nuclei and glandular lumens. Based on this, various quantitative indicators, including cell nuclear distribution, volume, and surface area, could be extracted. >
This research, with Juyeon Park, a student of the Integrated Master’s and Ph.D. Program at KAIST, as the first author, was published online in the prestigious journal Nature Communications on May 22.
(Paper title: Revealing 3D microanatomical structures of unlabeled thick cancer tissues using holotomography and virtual H&E staining.
[https://doi.org/10.1038/s41467-025-59820-0]
This study was supported by the Leader Researcher Program of the National Research Foundation of Korea, the Global Industry Technology Cooperation Center Project of the Korea Institute for Advancement of Technology, and the Korea Health Industry Development Institute.
2025.05.26 View 2598
KAIST Develops Virtual Staining Technology for 3D Histopathology
Moving beyond traditional methods of observing thinly sliced and stained cancer tissues, a collaborative international research team led by KAIST has successfully developed a groundbreaking technology. This innovation uses advanced optical techniques combined with an artificial intelligence-based deep learning algorithm to create realistic, virtually stained 3D images of cancer tissue without the need for serial sectioning nor staining. This breakthrough is anticipated to pave the way for next-generation non-invasive pathological diagnosis.
< Photo 1. (From left) Juyeon Park (Ph.D. Candidate, Department of Physics), Professor YongKeun Park (Department of Physics) (Top left) Professor Su-Jin Shin (Gangnam Severance Hospital), Professor Tae Hyun Hwang (Vanderbilt University School of Medicine) >
KAIST (President Kwang Hyung Lee) announced on the 26th that a research team led by Professor YongKeun Park of the Department of Physics, in collaboration with Professor Su-Jin Shin's team at Yonsei University Gangnam Severance Hospital, Professor Tae Hyun Hwang's team at Mayo Clinic, and Tomocube's AI research team, has developed an innovative technology capable of vividly displaying the 3D structure of cancer tissues without separate staining.
For over 200 years, conventional pathology has relied on observing cancer tissues under a microscope, a method that only shows specific cross-sections of the 3D cancer tissue. This has limited the ability to understand the three-dimensional connections and spatial arrangements between cells.
To overcome this, the research team utilized holotomography (HT), an advanced optical technology, to measure the 3D refractive index information of tissues. They then integrated an AI-based deep learning algorithm to successfully generate virtual H&E* images.* H&E (Hematoxylin & Eosin): The most widely used staining method for observing pathological tissues. Hematoxylin stains cell nuclei blue, and eosin stains cytoplasm pink.
The research team quantitatively demonstrated that the images generated by this technology are highly similar to actual stained tissue images. Furthermore, the technology exhibited consistent performance across various organs and tissues, proving its versatility and reliability as a next-generation pathological analysis tool.
< Figure 1. Comparison of conventional 3D tissue pathology procedure and the 3D virtual H&E staining technology proposed in this study. The traditional method requires preparing and staining dozens of tissue slides, while the proposed technology can reduce the number of slides by up to 10 times and quickly generate H&E images without the staining process. >
Moreover, by validating the feasibility of this technology through joint research with hospitals and research institutions in Korea and the United States, utilizing Tomocube's holotomography equipment, the team demonstrated its potential for full-scale adoption in real-world pathological research settings.
Professor YongKeun Park stated, "This research marks a major advancement by transitioning pathological analysis from conventional 2D methods to comprehensive 3D imaging. It will greatly enhance biomedical research and clinical diagnostics, particularly in understanding cancer tumor boundaries and the intricate spatial arrangements of cells within tumor microenvironments."
< Figure 2. Results of AI-based 3D virtual H&E staining and quantitative analysis of pathological tissue. The virtually stained images enabled 3D reconstruction of key pathological features such as cell nuclei and glandular lumens. Based on this, various quantitative indicators, including cell nuclear distribution, volume, and surface area, could be extracted. >
This research, with Juyeon Park, a student of the Integrated Master’s and Ph.D. Program at KAIST, as the first author, was published online in the prestigious journal Nature Communications on May 22.
(Paper title: Revealing 3D microanatomical structures of unlabeled thick cancer tissues using holotomography and virtual H&E staining.
[https://doi.org/10.1038/s41467-025-59820-0]
This study was supported by the Leader Researcher Program of the National Research Foundation of Korea, the Global Industry Technology Cooperation Center Project of the Korea Institute for Advancement of Technology, and the Korea Health Industry Development Institute.
2025.05.26 View 2598 -
 KAIST Succeeds in the Real-time Observation of Organoids using Holotomography
Organoids, which are 3D miniature organs that mimic the structure and function of human organs, play an essential role in disease research and drug development. A Korean research team has overcome the limitations of existing imaging technologies, succeeding in the real-time, high-resolution observation of living organoids.
KAIST (represented by President Kwang Hyung Lee) announced on the 14th of October that Professor YongKeun Park’s research team from the Department of Physics, in collaboration with the Genome Editing Research Center (Director Bon-Kyoung Koo) of the Institute for Basic Science (IBS President Do-Young Noh) and Tomocube Inc., has developed an imaging technology using holotomography to observe live, small intestinal organoids in real time at a high resolution.
Existing imaging techniques have struggled to observe living organoids in high resolution over extended periods and often required additional treatments like fluorescent staining.
< Figure 1. Overview of the low-coherence HT workflow. Using holotomography, 3D morphological restoration and quantitative analysis of organoids can be performed. In order to improve the limited field of view, which is a limitation of the microscope, our research team utilized a large-area field of view combination algorithm and made a 3D restoration by acquiring multi-focus holographic images for 3D measurements. After that, the organoids were compartmentalized to divide the parts necessary for analysis and quantitatively evaluated the protein concentration measurable from the refractive index and the survival rate of the organoids. >
The research team introduced holotomography technology to address these issues, which provides high-resolution images without the need for fluorescent staining and allows for the long-term observation of dynamic changes in real time without causing cell damage.
The team validated this technology using small intestinal organoids from experimental mice and were able to observe various cell structures inside the organoids in detail. They also captured dynamic changes such as growth processes, cell division, and cell death in real time using holotomography.
Additionally, the technology allowed for the precise analysis of the organoids' responses to drug treatments, verifying the survival of the cells.
The researchers believe that this breakthrough will open new horizons in organoid research, enabling the greater utilization of organoids in drug development, personalized medicine, and regenerative medicine.
Future research is expected to more accurately replicate the in vivo environment of organoids, contributing significantly to a more detailed understanding of various life phenomena at the cellular level through more precise 3D imaging.
< Figure 2. Real-time organoid morphology analysis. Using holotomography, it is possible to observe the lumen and villus development process of intestinal organoids in real time, which was difficult to observe with a conventional microscope. In addition, various information about intestinal organoids can be obtained by quantifying the size and protein amount of intestinal organoids through image analysis. >
Dr. Mahn Jae Lee, a graduate of KAIST's Graduate School of Medical Science and Engineering, currently at Chungnam National University Hospital and the first author of the paper, commented, "This research represents a new imaging technology that surpasses previous limitations and is expected to make a major contribution to disease modeling, personalized treatments, and drug development research using organoids."
The research results were published online in the international journal Experimental & Molecular Medicine on October 1, 2024, and the technology has been recognized for its applicability in various fields of life sciences. (Paper title: “Long-term three-dimensional high-resolution imaging of live unlabeled small intestinal organoids via low-coherence holotomography”)
This research was supported by the National Research Foundation of Korea, KAIST Institutes, and the Institute for Basic Science.
2024.10.14 View 5070
KAIST Succeeds in the Real-time Observation of Organoids using Holotomography
Organoids, which are 3D miniature organs that mimic the structure and function of human organs, play an essential role in disease research and drug development. A Korean research team has overcome the limitations of existing imaging technologies, succeeding in the real-time, high-resolution observation of living organoids.
KAIST (represented by President Kwang Hyung Lee) announced on the 14th of October that Professor YongKeun Park’s research team from the Department of Physics, in collaboration with the Genome Editing Research Center (Director Bon-Kyoung Koo) of the Institute for Basic Science (IBS President Do-Young Noh) and Tomocube Inc., has developed an imaging technology using holotomography to observe live, small intestinal organoids in real time at a high resolution.
Existing imaging techniques have struggled to observe living organoids in high resolution over extended periods and often required additional treatments like fluorescent staining.
< Figure 1. Overview of the low-coherence HT workflow. Using holotomography, 3D morphological restoration and quantitative analysis of organoids can be performed. In order to improve the limited field of view, which is a limitation of the microscope, our research team utilized a large-area field of view combination algorithm and made a 3D restoration by acquiring multi-focus holographic images for 3D measurements. After that, the organoids were compartmentalized to divide the parts necessary for analysis and quantitatively evaluated the protein concentration measurable from the refractive index and the survival rate of the organoids. >
The research team introduced holotomography technology to address these issues, which provides high-resolution images without the need for fluorescent staining and allows for the long-term observation of dynamic changes in real time without causing cell damage.
The team validated this technology using small intestinal organoids from experimental mice and were able to observe various cell structures inside the organoids in detail. They also captured dynamic changes such as growth processes, cell division, and cell death in real time using holotomography.
Additionally, the technology allowed for the precise analysis of the organoids' responses to drug treatments, verifying the survival of the cells.
The researchers believe that this breakthrough will open new horizons in organoid research, enabling the greater utilization of organoids in drug development, personalized medicine, and regenerative medicine.
Future research is expected to more accurately replicate the in vivo environment of organoids, contributing significantly to a more detailed understanding of various life phenomena at the cellular level through more precise 3D imaging.
< Figure 2. Real-time organoid morphology analysis. Using holotomography, it is possible to observe the lumen and villus development process of intestinal organoids in real time, which was difficult to observe with a conventional microscope. In addition, various information about intestinal organoids can be obtained by quantifying the size and protein amount of intestinal organoids through image analysis. >
Dr. Mahn Jae Lee, a graduate of KAIST's Graduate School of Medical Science and Engineering, currently at Chungnam National University Hospital and the first author of the paper, commented, "This research represents a new imaging technology that surpasses previous limitations and is expected to make a major contribution to disease modeling, personalized treatments, and drug development research using organoids."
The research results were published online in the international journal Experimental & Molecular Medicine on October 1, 2024, and the technology has been recognized for its applicability in various fields of life sciences. (Paper title: “Long-term three-dimensional high-resolution imaging of live unlabeled small intestinal organoids via low-coherence holotomography”)
This research was supported by the National Research Foundation of Korea, KAIST Institutes, and the Institute for Basic Science.
2024.10.14 View 5070 -
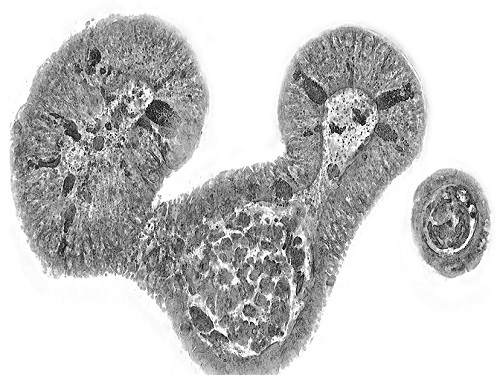 KAIST presents strategies for Holotomography in advanced bio research
Measuring and analyzing three-dimensional (3D) images of live cells and tissues is considered crucial in advanced fields of biology and medicine. Organoids, which are 3D structures that mimic organs, are particular examples that significantly benefits 3D live imaging. Organoids provide effective alternatives to animal testing in the drug development processes, and can rapidly determine personalized medicine. On the other hand, active researches are ongoing to utilize organoids for organ replacement.
< Figure 1. Schematic illustration of holotomography compared to X-ray CT. Similar to CT, they share the commonality of measuring the optical properties of an unlabeled specimen in three dimensions. Instead of X-rays, holotomography irradiates light in the visible range, and provides refractive index measurements of transparent specimens rather than absorptivity. While CT obtains three-dimensional information only through mechanical rotation of the irradiating light, holotomography can replace this by applying wavefront control technology in the visible range. >
Organelle-level observation of 3D biological specimens such as organoids and stem cell colonies without staining or preprocessing holds significant implications for both innovating basic research and bioindustrial applications related to regenerative medicine and bioindustrial applications.
Holotomography (HT) is a 3D optical microscopy that implements 3D reconstruction analogous to that of X-ray computed tomography (CT). Although HT and CT share a similar theoretical background, HT facilitates high-resolution examination inside cells and tissues, instead of the human body. HT obtains 3D images of cells and tissues at the organelle level without chemical or genetic labeling, thus overcomes various challenges of existing methods in bio research and industry. Its potential is highlighted in research fields where sample physiology must not be disrupted, such as regenerative medicine, personalized medicine, and infertility treatment.
< Figure 2. Label-free 3D imaging of diverse live cells. Time-lapse image of Hep3B cells illustrating subcellular morphology changes upon H2O2 treatment, followed by cellular recovery after returning to the regular cell culture medium. >
This paper introduces the advantages and broad applicability of HT to biomedical researchers, while presenting an overview of principles and future technical challenges to optical researchers. It showcases various cases of applying HT in studies such as 3D biology, regenerative medicine, and cancer research, as well as suggesting future optical development. Also, it categorizes HT based on the light source, to describe the principles, limitations, and improvements of each category in detail. Particularly, the paper addresses strategies for deepening cell and organoid studies by introducing artificial intelligence (AI) to HT.
Due to its potential to drive advanced bioindustry, HT is attracting interest and investment from universities and corporates worldwide. The KAIST research team has been leading this international field by developing core technologies and carrying out key application researches throughout the last decade.
< Figure 3. Various types of cells and organelles that make up the imaging barrier of a living intestinal organoid can be observed using holotomography. >
This paper, co-authored by Dr. Geon Kim from KAIST Research Center for Natural Sciences, Professor Ki-Jun Yoon's team from the Department of Biological Sciences, Director Bon-Kyoung Koo's team from the Institute for Basic Science (IBS) Center for Genome Engineering, and Dr. Seongsoo Lee's team from the Korea Basic Science Institute (KBSI), was published in 'Nature Reviews Methods Primers' on the 25th of July. This research was supported by the Leader Grant and Basic Science Research Program of the National Research Foundation, the Hologram Core Technology Development Grant of the Ministry of Science and ICT, the Nano and Material Technology Development Project, and the Health and Medical R&D Project of the Ministry of Health and Welfare.
2024.07.30 View 5590
KAIST presents strategies for Holotomography in advanced bio research
Measuring and analyzing three-dimensional (3D) images of live cells and tissues is considered crucial in advanced fields of biology and medicine. Organoids, which are 3D structures that mimic organs, are particular examples that significantly benefits 3D live imaging. Organoids provide effective alternatives to animal testing in the drug development processes, and can rapidly determine personalized medicine. On the other hand, active researches are ongoing to utilize organoids for organ replacement.
< Figure 1. Schematic illustration of holotomography compared to X-ray CT. Similar to CT, they share the commonality of measuring the optical properties of an unlabeled specimen in three dimensions. Instead of X-rays, holotomography irradiates light in the visible range, and provides refractive index measurements of transparent specimens rather than absorptivity. While CT obtains three-dimensional information only through mechanical rotation of the irradiating light, holotomography can replace this by applying wavefront control technology in the visible range. >
Organelle-level observation of 3D biological specimens such as organoids and stem cell colonies without staining or preprocessing holds significant implications for both innovating basic research and bioindustrial applications related to regenerative medicine and bioindustrial applications.
Holotomography (HT) is a 3D optical microscopy that implements 3D reconstruction analogous to that of X-ray computed tomography (CT). Although HT and CT share a similar theoretical background, HT facilitates high-resolution examination inside cells and tissues, instead of the human body. HT obtains 3D images of cells and tissues at the organelle level without chemical or genetic labeling, thus overcomes various challenges of existing methods in bio research and industry. Its potential is highlighted in research fields where sample physiology must not be disrupted, such as regenerative medicine, personalized medicine, and infertility treatment.
< Figure 2. Label-free 3D imaging of diverse live cells. Time-lapse image of Hep3B cells illustrating subcellular morphology changes upon H2O2 treatment, followed by cellular recovery after returning to the regular cell culture medium. >
This paper introduces the advantages and broad applicability of HT to biomedical researchers, while presenting an overview of principles and future technical challenges to optical researchers. It showcases various cases of applying HT in studies such as 3D biology, regenerative medicine, and cancer research, as well as suggesting future optical development. Also, it categorizes HT based on the light source, to describe the principles, limitations, and improvements of each category in detail. Particularly, the paper addresses strategies for deepening cell and organoid studies by introducing artificial intelligence (AI) to HT.
Due to its potential to drive advanced bioindustry, HT is attracting interest and investment from universities and corporates worldwide. The KAIST research team has been leading this international field by developing core technologies and carrying out key application researches throughout the last decade.
< Figure 3. Various types of cells and organelles that make up the imaging barrier of a living intestinal organoid can be observed using holotomography. >
This paper, co-authored by Dr. Geon Kim from KAIST Research Center for Natural Sciences, Professor Ki-Jun Yoon's team from the Department of Biological Sciences, Director Bon-Kyoung Koo's team from the Institute for Basic Science (IBS) Center for Genome Engineering, and Dr. Seongsoo Lee's team from the Korea Basic Science Institute (KBSI), was published in 'Nature Reviews Methods Primers' on the 25th of July. This research was supported by the Leader Grant and Basic Science Research Program of the National Research Foundation, the Hologram Core Technology Development Grant of the Ministry of Science and ICT, the Nano and Material Technology Development Project, and the Health and Medical R&D Project of the Ministry of Health and Welfare.
2024.07.30 View 5590 -
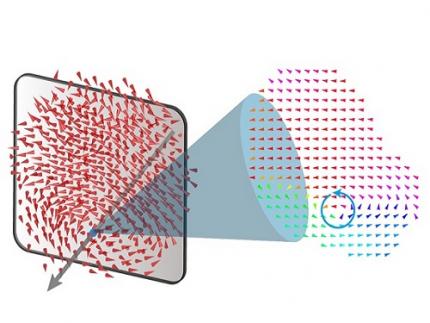 A 20-year-old puzzle solved: KAIST research team reveals the 'three-dimensional vortex' of zero-dimensional ferroelectrics
Materials that can maintain a magnetized state by themselves without an external magnetic field (i.e., permanent magnets) are called ferromagnets. Ferroelectrics can be thought of as the electric counterpart to ferromagnets, as they maintain a polarized state without an external electric field. It is well-known that ferromagnets lose their magnetic properties when reduced to nano sizes below a certain threshold. What happens when ferroelectrics are similarly made extremely small in all directions (i.e., into a zero-dimensional structure such as nanoparticles) has been a topic of controversy for a long time.
< (From left) Professor Yongsoo Yang, the corresponding author, and Chaehwa Jeong, the first author studying in the integrated master’s and doctoral program, of the KAIST Department of Physics >
The research team led by Dr. Yongsoo Yang from the Department of Physics at KAIST has, for the first time, experimentally clarified the three-dimensional, vortex-shaped polarization distribution inside ferroelectric nanoparticles through international collaborative research with POSTECH, SNU, KBSI, LBNL and University of Arkansas.
About 20 years ago, Prof. Laurent Bellaiche (currently at University of Arkansas) and his colleagues theoretically predicted that a unique form of polarization distribution, arranged in a toroidal vortex shape, could occur inside ferroelectric nanodots. They also suggested that if this vortex distribution could be properly controlled, it could be applied to ultra-high-density memory devices with capacities over 10,000 times greater than existing ones. However, experimental clarification had not been achieved due to the difficulty of measuring the three-dimensional polarization distribution within ferroelectric nanostructures.
The research team at KAIST successfully solved this 20-year-old challenge by implementing a technique called atomic electron tomography. This technique works by acquiring atomic-resolution transmission electron microscope images of the nanomaterials from multiple tilt angles, and then reconstructing them back into three-dimensional structures using advanced reconstruction algorithms. Electron tomography can be understood as essentially the same method with the CT scans used in hospitals to view internal organs in three dimensions; the KAIST team adapted it uniquely for nanomaterials, utilizing an electron microscope at the single-atom level.
< Figure 1. Three-dimensional polarization distribution of BaTiO3 nanoparticles revealed by atomic electron tomography. >(Left) Schematic of the electron tomography technique, which involves acquiring transmission electron microscope images at multiple tilt angles and reconstructing them into 3D atomic structures.(Center) Experimentally determined three-dimensional polarization distribution inside a BaTiO3 nanoparticle via atomic electron tomography. A vortex-like structure is clearly visible near the bottom (blue dot).(Right) A two-dimensional cross-section of the polarization distribution, thinly sliced at the center of the vortex, with the color and arrows together indicating the direction of the polarization. A distinct vortex structure can be observed.
Using atomic electron tomography, the team completely measured the positions of cation atoms inside barium titanate (BaTiO3) nanoparticles, a well-known ferroelectric material, in three dimensions. From the precisely determined 3D atomic arrangements, they were able to further calculate the internal three-dimensional polarization distribution at the single-atom level. The analysis of the polarization distribution revealed, for the first time experimentally, that topological polarization orderings including vortices, anti-vortices, skyrmions, and a Bloch point occur inside the 0-dimensional ferroelectrics, as theoretically predicted 20 years ago. Furthermore, it was also found that the number of internal vortices can be controlled depending on their sizes.
Prof. Sergey Prosandeev and Prof. Bellaiche (who proposed with other co-workers the polar vortex ordering theoretically 20 years ago), joined this collaboration and further proved that the vortex distribution results obtained from experiments are consistent with theoretical calculations.
By controlling the number and orientation of these polarization distributions, it is expected that this can be utilized into next-generation high-density memory device that can store more than 10,000 times the amount of information in the same-sized device compared to existing ones.
Dr. Yang, who led the research, explained the significance of the results: “This result suggests that controlling the size and shape of ferroelectrics alone, without needing to tune the substrate or surrounding environmental effects such as epitaxial strain, can manipulate ferroelectric vortices or other topological orderings at the nano-scale. Further research could then be applied to the development of next-generation ultra-high-density memory.”
This research, with Chaehwa Jeong from the Department of Physics at KAIST as the first author, was published online in Nature Communications on May 8th (Title: Revealing the Three-Dimensional Arrangement of Polar Topology in Nanoparticles).
The study was mainly supported by the National Research Foundation of Korea (NRF) Grants funded by the Korean Government (MSIT).
2024.05.31 View 7641
A 20-year-old puzzle solved: KAIST research team reveals the 'three-dimensional vortex' of zero-dimensional ferroelectrics
Materials that can maintain a magnetized state by themselves without an external magnetic field (i.e., permanent magnets) are called ferromagnets. Ferroelectrics can be thought of as the electric counterpart to ferromagnets, as they maintain a polarized state without an external electric field. It is well-known that ferromagnets lose their magnetic properties when reduced to nano sizes below a certain threshold. What happens when ferroelectrics are similarly made extremely small in all directions (i.e., into a zero-dimensional structure such as nanoparticles) has been a topic of controversy for a long time.
< (From left) Professor Yongsoo Yang, the corresponding author, and Chaehwa Jeong, the first author studying in the integrated master’s and doctoral program, of the KAIST Department of Physics >
The research team led by Dr. Yongsoo Yang from the Department of Physics at KAIST has, for the first time, experimentally clarified the three-dimensional, vortex-shaped polarization distribution inside ferroelectric nanoparticles through international collaborative research with POSTECH, SNU, KBSI, LBNL and University of Arkansas.
About 20 years ago, Prof. Laurent Bellaiche (currently at University of Arkansas) and his colleagues theoretically predicted that a unique form of polarization distribution, arranged in a toroidal vortex shape, could occur inside ferroelectric nanodots. They also suggested that if this vortex distribution could be properly controlled, it could be applied to ultra-high-density memory devices with capacities over 10,000 times greater than existing ones. However, experimental clarification had not been achieved due to the difficulty of measuring the three-dimensional polarization distribution within ferroelectric nanostructures.
The research team at KAIST successfully solved this 20-year-old challenge by implementing a technique called atomic electron tomography. This technique works by acquiring atomic-resolution transmission electron microscope images of the nanomaterials from multiple tilt angles, and then reconstructing them back into three-dimensional structures using advanced reconstruction algorithms. Electron tomography can be understood as essentially the same method with the CT scans used in hospitals to view internal organs in three dimensions; the KAIST team adapted it uniquely for nanomaterials, utilizing an electron microscope at the single-atom level.
< Figure 1. Three-dimensional polarization distribution of BaTiO3 nanoparticles revealed by atomic electron tomography. >(Left) Schematic of the electron tomography technique, which involves acquiring transmission electron microscope images at multiple tilt angles and reconstructing them into 3D atomic structures.(Center) Experimentally determined three-dimensional polarization distribution inside a BaTiO3 nanoparticle via atomic electron tomography. A vortex-like structure is clearly visible near the bottom (blue dot).(Right) A two-dimensional cross-section of the polarization distribution, thinly sliced at the center of the vortex, with the color and arrows together indicating the direction of the polarization. A distinct vortex structure can be observed.
Using atomic electron tomography, the team completely measured the positions of cation atoms inside barium titanate (BaTiO3) nanoparticles, a well-known ferroelectric material, in three dimensions. From the precisely determined 3D atomic arrangements, they were able to further calculate the internal three-dimensional polarization distribution at the single-atom level. The analysis of the polarization distribution revealed, for the first time experimentally, that topological polarization orderings including vortices, anti-vortices, skyrmions, and a Bloch point occur inside the 0-dimensional ferroelectrics, as theoretically predicted 20 years ago. Furthermore, it was also found that the number of internal vortices can be controlled depending on their sizes.
Prof. Sergey Prosandeev and Prof. Bellaiche (who proposed with other co-workers the polar vortex ordering theoretically 20 years ago), joined this collaboration and further proved that the vortex distribution results obtained from experiments are consistent with theoretical calculations.
By controlling the number and orientation of these polarization distributions, it is expected that this can be utilized into next-generation high-density memory device that can store more than 10,000 times the amount of information in the same-sized device compared to existing ones.
Dr. Yang, who led the research, explained the significance of the results: “This result suggests that controlling the size and shape of ferroelectrics alone, without needing to tune the substrate or surrounding environmental effects such as epitaxial strain, can manipulate ferroelectric vortices or other topological orderings at the nano-scale. Further research could then be applied to the development of next-generation ultra-high-density memory.”
This research, with Chaehwa Jeong from the Department of Physics at KAIST as the first author, was published online in Nature Communications on May 8th (Title: Revealing the Three-Dimensional Arrangement of Polar Topology in Nanoparticles).
The study was mainly supported by the National Research Foundation of Korea (NRF) Grants funded by the Korean Government (MSIT).
2024.05.31 View 7641 -
 Tomographic Measurement of Dielectric Tensors
Dielectric tensor tomography allows the direct measurement of the 3D dielectric tensors of optically anisotropic structures
A research team reported the direct measurement of dielectric tensors of anisotropic structures including the spatial variations of principal refractive indices and directors. The group also demonstrated quantitative tomographic measurements of various nematic liquid-crystal structures and their fast 3D nonequilibrium dynamics using a 3D label-free tomographic method. The method was described in Nature Materials.
Light-matter interactions are described by the dielectric tensor. Despite their importance in basic science and applications, it has not been possible to measure 3D dielectric tensors directly. The main challenge was due to the vectorial nature of light scattering from a 3D anisotropic structure. Previous approaches only addressed 3D anisotropic information indirectly and were limited to two-dimensional, qualitative, strict sample conditions or assumptions.
The research team developed a method enabling the tomographic reconstruction of 3D dielectric tensors without any preparation or assumptions. A sample is illuminated with a laser beam with various angles and circularly polarization states. Then, the light fields scattered from a sample are holographically measured and converted into vectorial diffraction components. Finally, by inversely solving a vectorial wave equation, the 3D dielectric tensor is reconstructed.
Professor YongKeun Park said, “There were a greater number of unknowns in direct measuring than with the conventional approach. We applied our approach to measure additional holographic images by slightly tilting the incident angle.”
He said that the slightly tilted illumination provides an additional orthogonal polarization, which makes the underdetermined problem become the determined problem. “Although scattered fields are dependent on the illumination angle, the Fourier differentiation theorem enables the extraction of the same dielectric tensor for the slightly tilted illumination,” Professor Park added.
His team’s method was validated by reconstructing well-known liquid crystal (LC) structures, including the twisted nematic, hybrid aligned nematic, radial, and bipolar configurations. Furthermore, the research team demonstrated the experimental measurements of the non-equilibrium dynamics of annihilating, nucleating, and merging LC droplets, and the LC polymer network with repeating 3D topological defects.
“This is the first experimental measurement of non-equilibrium dynamics and 3D topological defects in LC structures in a label-free manner. Our method enables the exploration of inaccessible nematic structures and interactions in non-equilibrium dynamics,” first author Dr. Seungwoo Shin explained.
-PublicationSeungwoo Shin, Jonghee Eun, Sang Seok Lee, Changjae Lee, Herve Hugonnet, Dong Ki Yoon, Shin-Hyun Kim, Jongwoo Jeong, YongKeun Park, “Tomographic Measurement ofDielectric Tensors at Optical Frequency,” Nature Materials March 02, 2022 (https://doi.org/10/1038/s41563-022-01202-8)
-ProfileProfessor YongKeun ParkBiomedical Optics Laboratory (http://bmol.kaist.ac.kr)Department of PhysicsCollege of Natural SciencesKAIST
2022.03.22 View 9457
Tomographic Measurement of Dielectric Tensors
Dielectric tensor tomography allows the direct measurement of the 3D dielectric tensors of optically anisotropic structures
A research team reported the direct measurement of dielectric tensors of anisotropic structures including the spatial variations of principal refractive indices and directors. The group also demonstrated quantitative tomographic measurements of various nematic liquid-crystal structures and their fast 3D nonequilibrium dynamics using a 3D label-free tomographic method. The method was described in Nature Materials.
Light-matter interactions are described by the dielectric tensor. Despite their importance in basic science and applications, it has not been possible to measure 3D dielectric tensors directly. The main challenge was due to the vectorial nature of light scattering from a 3D anisotropic structure. Previous approaches only addressed 3D anisotropic information indirectly and were limited to two-dimensional, qualitative, strict sample conditions or assumptions.
The research team developed a method enabling the tomographic reconstruction of 3D dielectric tensors without any preparation or assumptions. A sample is illuminated with a laser beam with various angles and circularly polarization states. Then, the light fields scattered from a sample are holographically measured and converted into vectorial diffraction components. Finally, by inversely solving a vectorial wave equation, the 3D dielectric tensor is reconstructed.
Professor YongKeun Park said, “There were a greater number of unknowns in direct measuring than with the conventional approach. We applied our approach to measure additional holographic images by slightly tilting the incident angle.”
He said that the slightly tilted illumination provides an additional orthogonal polarization, which makes the underdetermined problem become the determined problem. “Although scattered fields are dependent on the illumination angle, the Fourier differentiation theorem enables the extraction of the same dielectric tensor for the slightly tilted illumination,” Professor Park added.
His team’s method was validated by reconstructing well-known liquid crystal (LC) structures, including the twisted nematic, hybrid aligned nematic, radial, and bipolar configurations. Furthermore, the research team demonstrated the experimental measurements of the non-equilibrium dynamics of annihilating, nucleating, and merging LC droplets, and the LC polymer network with repeating 3D topological defects.
“This is the first experimental measurement of non-equilibrium dynamics and 3D topological defects in LC structures in a label-free manner. Our method enables the exploration of inaccessible nematic structures and interactions in non-equilibrium dynamics,” first author Dr. Seungwoo Shin explained.
-PublicationSeungwoo Shin, Jonghee Eun, Sang Seok Lee, Changjae Lee, Herve Hugonnet, Dong Ki Yoon, Shin-Hyun Kim, Jongwoo Jeong, YongKeun Park, “Tomographic Measurement ofDielectric Tensors at Optical Frequency,” Nature Materials March 02, 2022 (https://doi.org/10/1038/s41563-022-01202-8)
-ProfileProfessor YongKeun ParkBiomedical Optics Laboratory (http://bmol.kaist.ac.kr)Department of PhysicsCollege of Natural SciencesKAIST
2022.03.22 View 9457 -
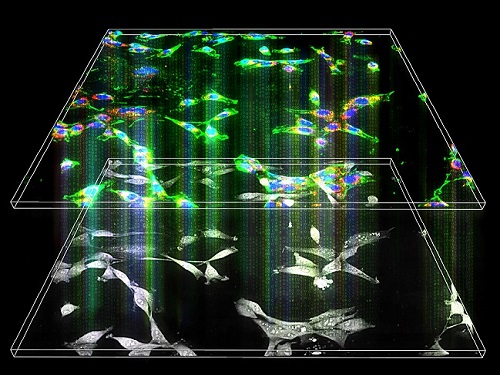 Label-Free Multiplexed Microtomography of Endogenous Subcellular Dynamics Using Deep Learning
AI-based holographic microscopy allows molecular imaging without introducing exogenous labeling agents
A research team upgraded the 3D microtomography observing dynamics of label-free live cells in multiplexed fluorescence imaging. The AI-powered 3D holotomographic microscopy extracts various molecular information from live unlabeled biological cells in real time without exogenous labeling or staining agents.
Professor YongKeum Park’s team and the startup Tomocube encoded 3D refractive index tomograms using the refractive index as a means of measurement. Then they decoded the information with a deep learning-based model that infers multiple 3D fluorescence tomograms from the refractive index measurements of the corresponding subcellular targets, thereby achieving multiplexed micro tomography. This study was reported in Nature Cell Biology online on December 7, 2021.
Fluorescence microscopy is the most widely used optical microscopy technique due to its high biochemical specificity. However, it needs to genetically manipulate or to stain cells with fluorescent labels in order to express fluorescent proteins. These labeling processes inevitably affect the intrinsic physiology of cells. It also has challenges in long-term measuring due to photobleaching and phototoxicity. The overlapped spectra of multiplexed fluorescence signals also hinder the viewing of various structures at the same time. More critically, it took several hours to observe the cells after preparing them.
3D holographic microscopy, also known as holotomography, is providing new ways to quantitatively image live cells without pretreatments such as staining. Holotomography can accurately and quickly measure the morphological and structural information of cells, but only provides limited biochemical and molecular information.
The 'AI microscope' created in this process takes advantage of the features of both holographic microscopy and fluorescence microscopy. That is, a specific image from a fluorescence microscope can be obtained without a fluorescent label. Therefore, the microscope can observe many types of cellular structures in their natural state in 3D and at the same time as fast as one millisecond, and long-term measurements over several days are also possible.
The Tomocube-KAIST team showed that fluorescence images can be directly and precisely predicted from holotomographic images in various cells and conditions. Using the quantitative relationship between the spatial distribution of the refractive index found by AI and the major structures in cells, it was possible to decipher the spatial distribution of the refractive index. And surprisingly, it confirmed that this relationship is constant regardless of cell type.
Professor Park said, “We were able to develop a new concept microscope that combines the advantages of several microscopes with the multidisciplinary research of AI, optics, and biology. It will be immediately applicable for new types of cells not included in the existing data and is expected to be widely applicable for various biological and medical research.”
When comparing the molecular image information extracted by AI with the molecular image information physically obtained by fluorescence staining in 3D space, it showed a 97% or more conformity, which is a level that is difficult to distinguish with the naked eye.
“Compared to the sub-60% accuracy of the fluorescence information extracted from the model developed by the Google AI team, it showed significantly higher performance,” Professor Park added.
This work was supported by the KAIST Up program, the BK21+ program, Tomocube, the National Research Foundation of Korea, and the Ministry of Science and ICT, and the Ministry of Health & Welfare.
-Publication
Hyun-seok Min, Won-Do Heo, YongKeun Park, et al. “Label-free multiplexed microtomography of endogenous subcellular dynamics using generalizable deep learning,” Nature Cell Biology (doi.org/10.1038/s41556-021-00802-x) published online December 07 2021.
-Profile
Professor YongKeun Park
Biomedical Optics Laboratory
Department of Physics
KAIST
2022.02.09 View 12489
Label-Free Multiplexed Microtomography of Endogenous Subcellular Dynamics Using Deep Learning
AI-based holographic microscopy allows molecular imaging without introducing exogenous labeling agents
A research team upgraded the 3D microtomography observing dynamics of label-free live cells in multiplexed fluorescence imaging. The AI-powered 3D holotomographic microscopy extracts various molecular information from live unlabeled biological cells in real time without exogenous labeling or staining agents.
Professor YongKeum Park’s team and the startup Tomocube encoded 3D refractive index tomograms using the refractive index as a means of measurement. Then they decoded the information with a deep learning-based model that infers multiple 3D fluorescence tomograms from the refractive index measurements of the corresponding subcellular targets, thereby achieving multiplexed micro tomography. This study was reported in Nature Cell Biology online on December 7, 2021.
Fluorescence microscopy is the most widely used optical microscopy technique due to its high biochemical specificity. However, it needs to genetically manipulate or to stain cells with fluorescent labels in order to express fluorescent proteins. These labeling processes inevitably affect the intrinsic physiology of cells. It also has challenges in long-term measuring due to photobleaching and phototoxicity. The overlapped spectra of multiplexed fluorescence signals also hinder the viewing of various structures at the same time. More critically, it took several hours to observe the cells after preparing them.
3D holographic microscopy, also known as holotomography, is providing new ways to quantitatively image live cells without pretreatments such as staining. Holotomography can accurately and quickly measure the morphological and structural information of cells, but only provides limited biochemical and molecular information.
The 'AI microscope' created in this process takes advantage of the features of both holographic microscopy and fluorescence microscopy. That is, a specific image from a fluorescence microscope can be obtained without a fluorescent label. Therefore, the microscope can observe many types of cellular structures in their natural state in 3D and at the same time as fast as one millisecond, and long-term measurements over several days are also possible.
The Tomocube-KAIST team showed that fluorescence images can be directly and precisely predicted from holotomographic images in various cells and conditions. Using the quantitative relationship between the spatial distribution of the refractive index found by AI and the major structures in cells, it was possible to decipher the spatial distribution of the refractive index. And surprisingly, it confirmed that this relationship is constant regardless of cell type.
Professor Park said, “We were able to develop a new concept microscope that combines the advantages of several microscopes with the multidisciplinary research of AI, optics, and biology. It will be immediately applicable for new types of cells not included in the existing data and is expected to be widely applicable for various biological and medical research.”
When comparing the molecular image information extracted by AI with the molecular image information physically obtained by fluorescence staining in 3D space, it showed a 97% or more conformity, which is a level that is difficult to distinguish with the naked eye.
“Compared to the sub-60% accuracy of the fluorescence information extracted from the model developed by the Google AI team, it showed significantly higher performance,” Professor Park added.
This work was supported by the KAIST Up program, the BK21+ program, Tomocube, the National Research Foundation of Korea, and the Ministry of Science and ICT, and the Ministry of Health & Welfare.
-Publication
Hyun-seok Min, Won-Do Heo, YongKeun Park, et al. “Label-free multiplexed microtomography of endogenous subcellular dynamics using generalizable deep learning,” Nature Cell Biology (doi.org/10.1038/s41556-021-00802-x) published online December 07 2021.
-Profile
Professor YongKeun Park
Biomedical Optics Laboratory
Department of Physics
KAIST
2022.02.09 View 12489 -
 Observing Individual Atoms in 3D Nanomaterials and Their Surfaces
Atoms are the basic building blocks for all materials. To tailor functional properties, it is essential to accurately determine their atomic structures. KAIST researchers observed the 3D atomic structure of a nanoparticle at the atom level via neural network-assisted atomic electron tomography.
Using a platinum nanoparticle as a model system, a research team led by Professor Yongsoo Yang demonstrated that an atomicity-based deep learning approach can reliably identify the 3D surface atomic structure with a precision of 15 picometers (only about 1/3 of a hydrogen atom’s radius). The atomic displacement, strain, and facet analysis revealed that the surface atomic structure and strain are related to both the shape of the nanoparticle and the particle-substrate interface.
Combined with quantum mechanical calculations such as density functional theory, the ability to precisely identify surface atomic structure will serve as a powerful key for understanding catalytic performance and oxidation effect.
“We solved the problem of determining the 3D surface atomic structure of nanomaterials in a reliable manner. It has been difficult to accurately measure the surface atomic structures due to the ‘missing wedge problem’ in electron tomography, which arises from geometrical limitations, allowing only part of a full tomographic angular range to be measured. We resolved the problem using a deep learning-based approach,” explained Professor Yang.
The missing wedge problem results in elongation and ringing artifacts, negatively affecting the accuracy of the atomic structure determined from the tomogram, especially for identifying the surface structures. The missing wedge problem has been the main roadblock for the precise determination of the 3D surface atomic structures of nanomaterials.
The team used atomic electron tomography (AET), which is basically a very high-resolution CT scan for nanomaterials using transmission electron microscopes. AET allows individual atom level 3D atomic structural determination.
“The main idea behind this deep learning-based approach is atomicity—the fact that all matter is composed of atoms. This means that true atomic resolution electron tomogram should only contain sharp 3D atomic potentials convolved with the electron beam profile,” said Professor Yang.
“A deep neural network can be trained using simulated tomograms that suffer from missing wedges as inputs, and the ground truth 3D atomic volumes as targets. The trained deep learning network effectively augments the imperfect tomograms and removes the artifacts resulting from the missing wedge problem.”
The precision of 3D atomic structure can be enhanced by nearly 70% by applying the deep learning-based augmentation. The accuracy of surface atom identification was also significantly improved.
Structure-property relationships of functional nanomaterials, especially the ones that strongly depend on the surface structures, such as catalytic properties for fuel-cell applications, can now be revealed at one of the most fundamental scales: the atomic scale.
Professor Yang concluded, “We would like to fully map out the 3D atomic structure with higher precision and better elemental specificity. And not being limited to atomic structures, we aim to measure the physical, chemical, and functional properties of nanomaterials at the 3D atomic scale by further advancing electron tomography techniques.”
This research, reported at Nature Communications, was funded by the National Research Foundation of Korea and the KAIST Global Singularity Research M3I3 Project.
-Publication
Juhyeok Lee, Chaehwa Jeong & Yongsoo Yang
“Single-atom level determination of 3-dimensional surface atomic structure via neural network-assisted atomic electron tomography”
Nature Communications
-Profile
Professor Yongsoo Yang
Department of Physics
Multi-Dimensional Atomic Imaging Lab (MDAIL)
http://mdail.kaist.ac.kr
KAIST
2021.05.12 View 13753
Observing Individual Atoms in 3D Nanomaterials and Their Surfaces
Atoms are the basic building blocks for all materials. To tailor functional properties, it is essential to accurately determine their atomic structures. KAIST researchers observed the 3D atomic structure of a nanoparticle at the atom level via neural network-assisted atomic electron tomography.
Using a platinum nanoparticle as a model system, a research team led by Professor Yongsoo Yang demonstrated that an atomicity-based deep learning approach can reliably identify the 3D surface atomic structure with a precision of 15 picometers (only about 1/3 of a hydrogen atom’s radius). The atomic displacement, strain, and facet analysis revealed that the surface atomic structure and strain are related to both the shape of the nanoparticle and the particle-substrate interface.
Combined with quantum mechanical calculations such as density functional theory, the ability to precisely identify surface atomic structure will serve as a powerful key for understanding catalytic performance and oxidation effect.
“We solved the problem of determining the 3D surface atomic structure of nanomaterials in a reliable manner. It has been difficult to accurately measure the surface atomic structures due to the ‘missing wedge problem’ in electron tomography, which arises from geometrical limitations, allowing only part of a full tomographic angular range to be measured. We resolved the problem using a deep learning-based approach,” explained Professor Yang.
The missing wedge problem results in elongation and ringing artifacts, negatively affecting the accuracy of the atomic structure determined from the tomogram, especially for identifying the surface structures. The missing wedge problem has been the main roadblock for the precise determination of the 3D surface atomic structures of nanomaterials.
The team used atomic electron tomography (AET), which is basically a very high-resolution CT scan for nanomaterials using transmission electron microscopes. AET allows individual atom level 3D atomic structural determination.
“The main idea behind this deep learning-based approach is atomicity—the fact that all matter is composed of atoms. This means that true atomic resolution electron tomogram should only contain sharp 3D atomic potentials convolved with the electron beam profile,” said Professor Yang.
“A deep neural network can be trained using simulated tomograms that suffer from missing wedges as inputs, and the ground truth 3D atomic volumes as targets. The trained deep learning network effectively augments the imperfect tomograms and removes the artifacts resulting from the missing wedge problem.”
The precision of 3D atomic structure can be enhanced by nearly 70% by applying the deep learning-based augmentation. The accuracy of surface atom identification was also significantly improved.
Structure-property relationships of functional nanomaterials, especially the ones that strongly depend on the surface structures, such as catalytic properties for fuel-cell applications, can now be revealed at one of the most fundamental scales: the atomic scale.
Professor Yang concluded, “We would like to fully map out the 3D atomic structure with higher precision and better elemental specificity. And not being limited to atomic structures, we aim to measure the physical, chemical, and functional properties of nanomaterials at the 3D atomic scale by further advancing electron tomography techniques.”
This research, reported at Nature Communications, was funded by the National Research Foundation of Korea and the KAIST Global Singularity Research M3I3 Project.
-Publication
Juhyeok Lee, Chaehwa Jeong & Yongsoo Yang
“Single-atom level determination of 3-dimensional surface atomic structure via neural network-assisted atomic electron tomography”
Nature Communications
-Profile
Professor Yongsoo Yang
Department of Physics
Multi-Dimensional Atomic Imaging Lab (MDAIL)
http://mdail.kaist.ac.kr
KAIST
2021.05.12 View 13753 -
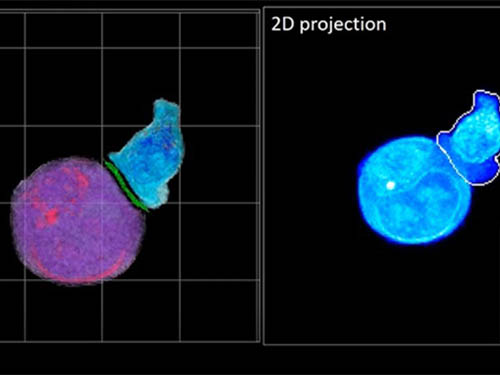 Deep-Learning and 3D Holographic Microscopy Beats Scientists at Analyzing Cancer Immunotherapy
Live tracking and analyzing of the dynamics of chimeric antigen receptor (CAR) T-cells targeting cancer cells can open new avenues for the development of cancer immunotherapy. However, imaging via conventional microscopy approaches can result in cellular damage, and assessments of cell-to-cell interactions are extremely difficult and labor-intensive. When researchers applied deep learning and 3D holographic microscopy to the task, however, they not only avoided these difficultues but found that AI was better at it than humans were.
Artificial intelligence (AI) is helping researchers decipher images from a new holographic microscopy technique needed to investigate a key process in cancer immunotherapy “live” as it takes place. The AI transformed work that, if performed manually by scientists, would otherwise be incredibly labor-intensive and time-consuming into one that is not only effortless but done better than they could have done it themselves. The research, conducted by the team of Professor YongKeun Park from the Department of Physics, appeared in the journal eLife last December.
A critical stage in the development of the human immune system’s ability to respond not just generally to any invader (such as pathogens or cancer cells) but specifically to that particular type of invader and remember it should it attempt to invade again is the formation of a junction between an immune cell called a T-cell and a cell that presents the antigen, or part of the invader that is causing the problem, to it. This process is like when a picture of a suspect is sent to a police car so that the officers can recognize the criminal they are trying to track down. The junction between the two cells, called the immunological synapse, or IS, is the key process in teaching the immune system how to recognize a specific type of invader.
Since the formation of the IS junction is such a critical step for the initiation of an antigen-specific immune response, various techniques allowing researchers to observe the process as it happens have been used to study its dynamics. Most of these live imaging techniques rely on fluorescence microscopy, where genetic tweaking causes part of a protein from a cell to fluoresce, in turn allowing the subject to be tracked via fluorescence rather than via the reflected light used in many conventional microscopy techniques.
However, fluorescence-based imaging can suffer from effects such as photo-bleaching and photo-toxicity, preventing the assessment of dynamic changes in the IS junction process over the long term. Fluorescence-based imaging still involves illumination, whereupon the fluorophores (chemical compounds that cause the fluorescence) emit light of a different color. Photo-bleaching or photo-toxicity occur when the subject is exposed to too much illumination, resulting in chemical alteration or cellular damage.
One recent option that does away with fluorescent labelling and thereby avoids such problems is 3D holographic microscopy or holotomography (HT). In this technique, the refractive index (the way that light changes direction when encountering a substance with a different density—why a straw looks like it bends in a glass of water) is recorded in 3D as a hologram.
Until now, HT has been used to study single cells, but never cell-cell interactions involved in immune responses. One of the main reasons is the difficulty of “segmentation,” or distinguishing the different parts of a cell and thus distinguishing between the interacting cells; in other words, deciphering which part belongs to which cell.
Manual segmentation, or marking out the different parts manually, is one option, but it is difficult and time-consuming, especially in three dimensions. To overcome this problem, automatic segmentation has been developed in which simple computer algorithms perform the identification.
“But these basic algorithms often make mistakes,” explained Professor YongKeun Park, “particularly with respect to adjoining segmentation, which of course is exactly what is occurring here in the immune response we’re most interested in.”
So, the researchers applied a deep learning framework to the HT segmentation problem. Deep learning is a type of machine learning in which artificial neural networks based on the human brain recognize patterns in a way that is similar to how humans do this. Regular machine learning requires data as an input that has already been labelled. The AI “learns” by understanding the labeled data and then recognizes the concept that has been labelled when it is fed novel data. For example, AI trained on a thousand images of cats labelled “cat” should be able to recognize a cat the next time it encounters an image with a cat in it. Deep learning involves multiple layers of artificial neural networks attacking much larger, but unlabeled datasets, in which the AI develops its own ‘labels’ for concepts it encounters.
In essence, the deep learning framework that KAIST researchers developed, called DeepIS, came up with its own concepts by which it distinguishes the different parts of the IS junction process. To validate this method, the research team applied it to the dynamics of a particular IS junction formed between chimeric antigen receptor (CAR) T-cells and target cancer cells. They then compared the results to what they would normally have done: the laborious process of performing the segmentation manually. They found not only that DeepIS was able to define areas within the IS with high accuracy, but that the technique was even able to capture information about the total distribution of proteins within the IS that may not have been easily measured using conventional techniques.
“In addition to allowing us to avoid the drudgery of manual segmentation and the problems of photo-bleaching and photo-toxicity, we found that the AI actually did a better job,” Professor Park added.
The next step will be to combine the technique with methods of measuring how much physical force is applied by different parts of the IS junction, such as holographic optical tweezers or traction force microscopy.
-Profile
Professor YongKeun Park
Department of Physics
Biomedical Optics Laboratory
http://bmol.kaist.ac.kr
KAIST
2021.02.24 View 13952
Deep-Learning and 3D Holographic Microscopy Beats Scientists at Analyzing Cancer Immunotherapy
Live tracking and analyzing of the dynamics of chimeric antigen receptor (CAR) T-cells targeting cancer cells can open new avenues for the development of cancer immunotherapy. However, imaging via conventional microscopy approaches can result in cellular damage, and assessments of cell-to-cell interactions are extremely difficult and labor-intensive. When researchers applied deep learning and 3D holographic microscopy to the task, however, they not only avoided these difficultues but found that AI was better at it than humans were.
Artificial intelligence (AI) is helping researchers decipher images from a new holographic microscopy technique needed to investigate a key process in cancer immunotherapy “live” as it takes place. The AI transformed work that, if performed manually by scientists, would otherwise be incredibly labor-intensive and time-consuming into one that is not only effortless but done better than they could have done it themselves. The research, conducted by the team of Professor YongKeun Park from the Department of Physics, appeared in the journal eLife last December.
A critical stage in the development of the human immune system’s ability to respond not just generally to any invader (such as pathogens or cancer cells) but specifically to that particular type of invader and remember it should it attempt to invade again is the formation of a junction between an immune cell called a T-cell and a cell that presents the antigen, or part of the invader that is causing the problem, to it. This process is like when a picture of a suspect is sent to a police car so that the officers can recognize the criminal they are trying to track down. The junction between the two cells, called the immunological synapse, or IS, is the key process in teaching the immune system how to recognize a specific type of invader.
Since the formation of the IS junction is such a critical step for the initiation of an antigen-specific immune response, various techniques allowing researchers to observe the process as it happens have been used to study its dynamics. Most of these live imaging techniques rely on fluorescence microscopy, where genetic tweaking causes part of a protein from a cell to fluoresce, in turn allowing the subject to be tracked via fluorescence rather than via the reflected light used in many conventional microscopy techniques.
However, fluorescence-based imaging can suffer from effects such as photo-bleaching and photo-toxicity, preventing the assessment of dynamic changes in the IS junction process over the long term. Fluorescence-based imaging still involves illumination, whereupon the fluorophores (chemical compounds that cause the fluorescence) emit light of a different color. Photo-bleaching or photo-toxicity occur when the subject is exposed to too much illumination, resulting in chemical alteration or cellular damage.
One recent option that does away with fluorescent labelling and thereby avoids such problems is 3D holographic microscopy or holotomography (HT). In this technique, the refractive index (the way that light changes direction when encountering a substance with a different density—why a straw looks like it bends in a glass of water) is recorded in 3D as a hologram.
Until now, HT has been used to study single cells, but never cell-cell interactions involved in immune responses. One of the main reasons is the difficulty of “segmentation,” or distinguishing the different parts of a cell and thus distinguishing between the interacting cells; in other words, deciphering which part belongs to which cell.
Manual segmentation, or marking out the different parts manually, is one option, but it is difficult and time-consuming, especially in three dimensions. To overcome this problem, automatic segmentation has been developed in which simple computer algorithms perform the identification.
“But these basic algorithms often make mistakes,” explained Professor YongKeun Park, “particularly with respect to adjoining segmentation, which of course is exactly what is occurring here in the immune response we’re most interested in.”
So, the researchers applied a deep learning framework to the HT segmentation problem. Deep learning is a type of machine learning in which artificial neural networks based on the human brain recognize patterns in a way that is similar to how humans do this. Regular machine learning requires data as an input that has already been labelled. The AI “learns” by understanding the labeled data and then recognizes the concept that has been labelled when it is fed novel data. For example, AI trained on a thousand images of cats labelled “cat” should be able to recognize a cat the next time it encounters an image with a cat in it. Deep learning involves multiple layers of artificial neural networks attacking much larger, but unlabeled datasets, in which the AI develops its own ‘labels’ for concepts it encounters.
In essence, the deep learning framework that KAIST researchers developed, called DeepIS, came up with its own concepts by which it distinguishes the different parts of the IS junction process. To validate this method, the research team applied it to the dynamics of a particular IS junction formed between chimeric antigen receptor (CAR) T-cells and target cancer cells. They then compared the results to what they would normally have done: the laborious process of performing the segmentation manually. They found not only that DeepIS was able to define areas within the IS with high accuracy, but that the technique was even able to capture information about the total distribution of proteins within the IS that may not have been easily measured using conventional techniques.
“In addition to allowing us to avoid the drudgery of manual segmentation and the problems of photo-bleaching and photo-toxicity, we found that the AI actually did a better job,” Professor Park added.
The next step will be to combine the technique with methods of measuring how much physical force is applied by different parts of the IS junction, such as holographic optical tweezers or traction force microscopy.
-Profile
Professor YongKeun Park
Department of Physics
Biomedical Optics Laboratory
http://bmol.kaist.ac.kr
KAIST
2021.02.24 View 13952 -
 Professor YongKeun Park Elected as a Fellow of the Optical Society
Professor YongKeun Park, from the Department of Physics at KAIST, was elected as a fellow member of the Optical Society (OSA) in Washington, D.C. on September 12. Fellow membership is given to members who have made a significant contribution to the advancement of optics and photonics.
Professor Park was recognized for his research on digital holography and wavefront control technology.
Professor Park has been producing outstanding research outcomes in the field of holographic technology and light scattering control since joining KAIST in 2010. In particular, he developed and commercialized technology for a holographic telescope. He applied it to various medical and biological research projects, leading the field worldwide.
In the past, cells needed to be dyed with fluorescent materials to capture a 3-D image. However, Professor Park’s holotomography (HT) technology can capture 3-D images of living cells and tissues in real time without color dyeing. This technology allows diversified research in the biological and medical field.
Professor Park established a company, Tomocube, Inc. in 2015 to commercialize the technology. In 2016, he received funding from SoftBank Ventures and Hanmi Pharmaceutical. Currently, major institutes, including MIT, the University of Pittsburgh, the German Cancer Research Center, and Seoul National University Hospital are using his equipment.
Recently, Professor Park and his team developed technology based on light scattering measurements. With this technology, they established a company called The Wave Talk and received funding from various organizations, such as NAVER. Its first product is about to be released.
Professor Park said, “I am glad to become a fellow member based on the research outcomes I produced since I was appointed as a professor at KAIST. I would like to thank the excellent researchers as well as the school for its support. I will devote myself to continuously producing novel outcomes in both basic and applied fields.”
Professor Park has published nearly 100 papers in renowned journals including Nature Photonics, Nature Communications, Science Advances, and Physical Review Letters.
2017.10.18 View 13874
Professor YongKeun Park Elected as a Fellow of the Optical Society
Professor YongKeun Park, from the Department of Physics at KAIST, was elected as a fellow member of the Optical Society (OSA) in Washington, D.C. on September 12. Fellow membership is given to members who have made a significant contribution to the advancement of optics and photonics.
Professor Park was recognized for his research on digital holography and wavefront control technology.
Professor Park has been producing outstanding research outcomes in the field of holographic technology and light scattering control since joining KAIST in 2010. In particular, he developed and commercialized technology for a holographic telescope. He applied it to various medical and biological research projects, leading the field worldwide.
In the past, cells needed to be dyed with fluorescent materials to capture a 3-D image. However, Professor Park’s holotomography (HT) technology can capture 3-D images of living cells and tissues in real time without color dyeing. This technology allows diversified research in the biological and medical field.
Professor Park established a company, Tomocube, Inc. in 2015 to commercialize the technology. In 2016, he received funding from SoftBank Ventures and Hanmi Pharmaceutical. Currently, major institutes, including MIT, the University of Pittsburgh, the German Cancer Research Center, and Seoul National University Hospital are using his equipment.
Recently, Professor Park and his team developed technology based on light scattering measurements. With this technology, they established a company called The Wave Talk and received funding from various organizations, such as NAVER. Its first product is about to be released.
Professor Park said, “I am glad to become a fellow member based on the research outcomes I produced since I was appointed as a professor at KAIST. I would like to thank the excellent researchers as well as the school for its support. I will devote myself to continuously producing novel outcomes in both basic and applied fields.”
Professor Park has published nearly 100 papers in renowned journals including Nature Photonics, Nature Communications, Science Advances, and Physical Review Letters.
2017.10.18 View 13874 -
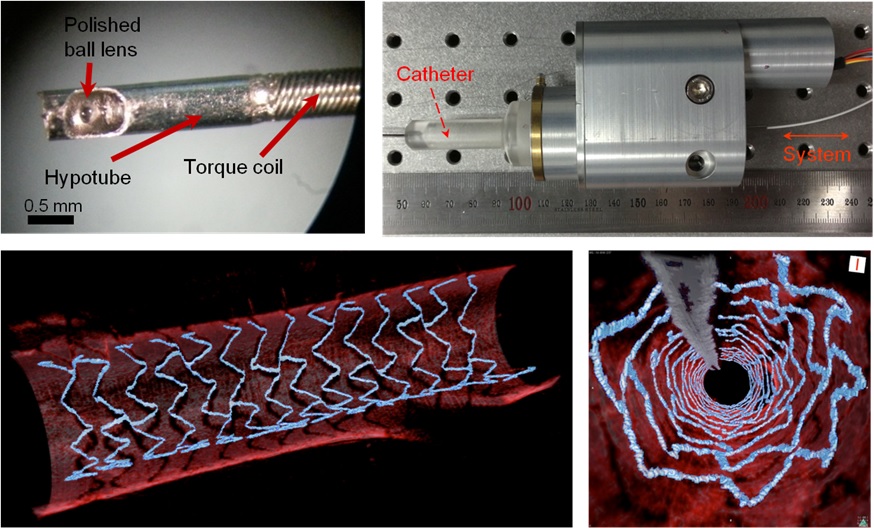 High Resolution 3D Blood Vessel Endoscope System Developed
Professor Wangyeol Oh of KAIST’s Mechanical Engineering Department has succeeded in developing an optical imaging endoscope system that employs an imaging velocity, which is up to 3.5 times faster than the previous systems. Furthermore, he has utilized this endoscope to acquire the world’s first high-resolution 3D images of the insides of in vivo blood vessel.
Professor Oh’s work is Korea’s first development of blood vessel endoscope system, possessing an imaging speed, resolution, imaging quality, and image-capture area. The system can also simultaneously perform a functional imaging, such as polarized imaging, which is advantageous for identifying the vulnerability of the blood vessel walls.
The Endoscopic Optical Coherence Tomography (OCT) System provides the highest resolution that is used to diagnose cardiovascular diseases, represented mainly by myocardial infarction.
However, the previous system was not fast enough to take images inside of the vessels, and therefore it was often impossible to accurately identify and analyze the vessel condition. To achieve an in vivo blood vessel optical imaging in clinical trials, the endoscope needed to be inserted, after which a clear liquid flows instantly, and pictures can be taken in only a few seconds.
The KAIST research team proposed a solution for such problem by developing a high-speed, high-resolution optical tomographic imaging system, a flexible endoscope with a diameter of 0.8 mm, as well as a device that can scan the imaging light within the blood vessels at high speed. Then, these devices were combined to visualize the internal structure of the vessel wall.
Using the developed system, the researchers were able to obtain high-resolution images of about 7 cm blood vessels of a rabbit’s aorta, which is similar size to human’s coronary arteries. The tomography scan took only 5.8 seconds, at a speed of 350 scans per second in all three directions with a resolution of 10~35㎛.
If the images are taken every 200 ㎛, like the currently available commercial vascular imaging endoscopes, a 7cm length vessel can be imaged in only one second.
Professor Wangyeol Oh said, “Our newly developed blood vessel endoscope system was tested by imaging a live animal’s blood vessels, which is similar to human blood vessels. The result was very successful.”
“Collaborating closely with hospitals, we are preparing to produce the imaging of an animal’s coronary arteries, which is similar in size to the human heart,” commented Professor Oh on the future clinical application and commercialization of the endoscope system. He added, “After such procedures, the technique can be applied in clinical patients within a few years.”
Professor Oh’s research was supported by the National Research Foundation of Korea and the Global Frontier Project by the Korean government. The research results were published in the 2014 January’s edition of Biomedical Optics Express.
Figure 1: End portion of optical endoscope (upper left)
Figure 2: High-speed optical scanning unit of the endoscope (top right)
Figure 3: High-resolution images of the inside of in vivo animal blood vessels (in the direction of vascular circumference and length)
Figure 4: High-resolution images of the inside of in vivo animal blood vessels (in the direction of the vein depth)
2014.03.25 View 13036
High Resolution 3D Blood Vessel Endoscope System Developed
Professor Wangyeol Oh of KAIST’s Mechanical Engineering Department has succeeded in developing an optical imaging endoscope system that employs an imaging velocity, which is up to 3.5 times faster than the previous systems. Furthermore, he has utilized this endoscope to acquire the world’s first high-resolution 3D images of the insides of in vivo blood vessel.
Professor Oh’s work is Korea’s first development of blood vessel endoscope system, possessing an imaging speed, resolution, imaging quality, and image-capture area. The system can also simultaneously perform a functional imaging, such as polarized imaging, which is advantageous for identifying the vulnerability of the blood vessel walls.
The Endoscopic Optical Coherence Tomography (OCT) System provides the highest resolution that is used to diagnose cardiovascular diseases, represented mainly by myocardial infarction.
However, the previous system was not fast enough to take images inside of the vessels, and therefore it was often impossible to accurately identify and analyze the vessel condition. To achieve an in vivo blood vessel optical imaging in clinical trials, the endoscope needed to be inserted, after which a clear liquid flows instantly, and pictures can be taken in only a few seconds.
The KAIST research team proposed a solution for such problem by developing a high-speed, high-resolution optical tomographic imaging system, a flexible endoscope with a diameter of 0.8 mm, as well as a device that can scan the imaging light within the blood vessels at high speed. Then, these devices were combined to visualize the internal structure of the vessel wall.
Using the developed system, the researchers were able to obtain high-resolution images of about 7 cm blood vessels of a rabbit’s aorta, which is similar size to human’s coronary arteries. The tomography scan took only 5.8 seconds, at a speed of 350 scans per second in all three directions with a resolution of 10~35㎛.
If the images are taken every 200 ㎛, like the currently available commercial vascular imaging endoscopes, a 7cm length vessel can be imaged in only one second.
Professor Wangyeol Oh said, “Our newly developed blood vessel endoscope system was tested by imaging a live animal’s blood vessels, which is similar to human blood vessels. The result was very successful.”
“Collaborating closely with hospitals, we are preparing to produce the imaging of an animal’s coronary arteries, which is similar in size to the human heart,” commented Professor Oh on the future clinical application and commercialization of the endoscope system. He added, “After such procedures, the technique can be applied in clinical patients within a few years.”
Professor Oh’s research was supported by the National Research Foundation of Korea and the Global Frontier Project by the Korean government. The research results were published in the 2014 January’s edition of Biomedical Optics Express.
Figure 1: End portion of optical endoscope (upper left)
Figure 2: High-speed optical scanning unit of the endoscope (top right)
Figure 3: High-resolution images of the inside of in vivo animal blood vessels (in the direction of vascular circumference and length)
Figure 4: High-resolution images of the inside of in vivo animal blood vessels (in the direction of the vein depth)
2014.03.25 View 13036 -
 The key to Alzheimer disease, PET-MRI made in Korea
Professor Kyu-Sung Cho
- Simultaneous PET-MRI imaging system commercialization technology developed purely from domestic technology - - Inspiring achievement by KAIST, National NanoFab Center, Sogang University, Seoul National University Hospital –
Hopes are high for the potential of producing domestic products in the field of state-of-the-art medical imaging equipment that used to rely on imported products.
The joint research team (KAIST, Sogang University and Seoul National University) with KAIST Department of Nuclear and Quantum Engineering Professor Kyu-Sung Cho in charge, together with National Nanofab Institution (NNFC; Director Jae-Young Lee), has developed PET-MRI simultaneous imaging system with domestic technology only. The team successfully acquired brain images of 3 volunteers with the newly developed system.
PET-MRI is integrated state-of-the-art medical imaging equipment that combines the advantages of Magnetic Resonance Imaging (MRI) that shows anatomical images of the body and Position Emission Tomography (PET) that analyses cell activity and metabolism. Since the anatomical information and functional information can be seen simultaneously, the device can be used to diagnose early onset Alzheimer’s disease and is essential in biological science research, such as new medicine development.
The existing equipment used to take MRI and PET images separately due to the strong magnetic field generated by MRI and combine the images. Hence, it was time consuming and error-prone due to patient’s movement. There was a need to develop PET that functions within a magnetic field to create a simultaneous imaging system.
The newly developed integral PET-MRI has 3 technical characteristics: 1. PET detector without magnetic interference, 2. PET-MRI integration system, 3.PET-MRI imaging processing.
The PET detector is the most important factor and accounts for half the cost of the whole system. KAIST Professor Cho and NNFC Doctor Woo-Suk Seol’s team successfully developed the Silicon Photomultiplier (amplifies light coming into the radiation detector) that can be used in strong magnetic fields. The developed sensor has a global competitive edge since it optimises semiconductor processing to yield over 95% productivity and around 10% gamma radiation energy resolving power.
Sogang University Department and Electrical Engineering Professor Yong Choi developed cutting edge PET system using a new concept of electric charge signal transmission method and imaging location distinction circuit. The creativity and excellence of the research findings were recognised and hence published on the cover of Medical Physics in June.
Seoul National University Hospital Department of Nuclear Medicine Professor Jae-Sung Lee developed the Silicon Photomultiplier sensor based PET imaging reconstitution programme, MRI imaging based PET imaging revision technology and PET-MRI imaging integration software.
Furthermore, KAIST Department of Electrical Engineering Professor Hyun-Wook Park was responsible for the development of RF Shielding technology that enables simultaneous installation of PET and MRI and using this technology, he developed a head coil for the brain that can be connected to PET for installation.
Based on the technology describe above, the joint research team successfully developed PET-MRI system for brains and acquired PET-MRI integrated brain images from 3 volunteers last June.
In particular, this system has the distinct feature of a detachable PET module and MRI head coil to the existing whole body MRI, so that PET-MRI simultaneous imaging is possible with low installation cost.
Professor Cho said, “We have prepared the foundation of domestic commercial PET and the system has a competitive edge in the global market of PET-MRI system technology.” He continued, “It can reduce the cost of the increasing brain related disease diagnosis, including Alzheimer’s, dramatically.”
Funded by Ministry of Trade, Industry and Energy as an Industrial Foundation Technology Development Project (98 billion won in 7 years), the research applied for over 20 patents and 20 CSI theses.
Figure 1.Brain phantom images from developed PET-MRI system
Figure 2. Brain images from developed PET-MRI system
Figure 3. Domestic PET-MRI clinical trial
Figure 4. Head RF coil and PET detector inserted in MRI
Figure 5. Insertion type PET detector module
Figure 6. Silicon Photomultiplier sensor (Left) and flash crystal block (right)
Figure7. Silicon Photomultiplier sensor
Figure 8. PET detection principle
2013.11.28 View 15343
The key to Alzheimer disease, PET-MRI made in Korea
Professor Kyu-Sung Cho
- Simultaneous PET-MRI imaging system commercialization technology developed purely from domestic technology - - Inspiring achievement by KAIST, National NanoFab Center, Sogang University, Seoul National University Hospital –
Hopes are high for the potential of producing domestic products in the field of state-of-the-art medical imaging equipment that used to rely on imported products.
The joint research team (KAIST, Sogang University and Seoul National University) with KAIST Department of Nuclear and Quantum Engineering Professor Kyu-Sung Cho in charge, together with National Nanofab Institution (NNFC; Director Jae-Young Lee), has developed PET-MRI simultaneous imaging system with domestic technology only. The team successfully acquired brain images of 3 volunteers with the newly developed system.
PET-MRI is integrated state-of-the-art medical imaging equipment that combines the advantages of Magnetic Resonance Imaging (MRI) that shows anatomical images of the body and Position Emission Tomography (PET) that analyses cell activity and metabolism. Since the anatomical information and functional information can be seen simultaneously, the device can be used to diagnose early onset Alzheimer’s disease and is essential in biological science research, such as new medicine development.
The existing equipment used to take MRI and PET images separately due to the strong magnetic field generated by MRI and combine the images. Hence, it was time consuming and error-prone due to patient’s movement. There was a need to develop PET that functions within a magnetic field to create a simultaneous imaging system.
The newly developed integral PET-MRI has 3 technical characteristics: 1. PET detector without magnetic interference, 2. PET-MRI integration system, 3.PET-MRI imaging processing.
The PET detector is the most important factor and accounts for half the cost of the whole system. KAIST Professor Cho and NNFC Doctor Woo-Suk Seol’s team successfully developed the Silicon Photomultiplier (amplifies light coming into the radiation detector) that can be used in strong magnetic fields. The developed sensor has a global competitive edge since it optimises semiconductor processing to yield over 95% productivity and around 10% gamma radiation energy resolving power.
Sogang University Department and Electrical Engineering Professor Yong Choi developed cutting edge PET system using a new concept of electric charge signal transmission method and imaging location distinction circuit. The creativity and excellence of the research findings were recognised and hence published on the cover of Medical Physics in June.
Seoul National University Hospital Department of Nuclear Medicine Professor Jae-Sung Lee developed the Silicon Photomultiplier sensor based PET imaging reconstitution programme, MRI imaging based PET imaging revision technology and PET-MRI imaging integration software.
Furthermore, KAIST Department of Electrical Engineering Professor Hyun-Wook Park was responsible for the development of RF Shielding technology that enables simultaneous installation of PET and MRI and using this technology, he developed a head coil for the brain that can be connected to PET for installation.
Based on the technology describe above, the joint research team successfully developed PET-MRI system for brains and acquired PET-MRI integrated brain images from 3 volunteers last June.
In particular, this system has the distinct feature of a detachable PET module and MRI head coil to the existing whole body MRI, so that PET-MRI simultaneous imaging is possible with low installation cost.
Professor Cho said, “We have prepared the foundation of domestic commercial PET and the system has a competitive edge in the global market of PET-MRI system technology.” He continued, “It can reduce the cost of the increasing brain related disease diagnosis, including Alzheimer’s, dramatically.”
Funded by Ministry of Trade, Industry and Energy as an Industrial Foundation Technology Development Project (98 billion won in 7 years), the research applied for over 20 patents and 20 CSI theses.
Figure 1.Brain phantom images from developed PET-MRI system
Figure 2. Brain images from developed PET-MRI system
Figure 3. Domestic PET-MRI clinical trial
Figure 4. Head RF coil and PET detector inserted in MRI
Figure 5. Insertion type PET detector module
Figure 6. Silicon Photomultiplier sensor (Left) and flash crystal block (right)
Figure7. Silicon Photomultiplier sensor
Figure 8. PET detection principle
2013.11.28 View 15343