BIO
-
 Tomographic Measurement of Dielectric Tensors
Dielectric tensor tomography allows the direct measurement of the 3D dielectric tensors of optically anisotropic structures
A research team reported the direct measurement of dielectric tensors of anisotropic structures including the spatial variations of principal refractive indices and directors. The group also demonstrated quantitative tomographic measurements of various nematic liquid-crystal structures and their fast 3D nonequilibrium dynamics using a 3D label-free tomographic method. The method was described in Nature Materials.
Light-matter interactions are described by the dielectric tensor. Despite their importance in basic science and applications, it has not been possible to measure 3D dielectric tensors directly. The main challenge was due to the vectorial nature of light scattering from a 3D anisotropic structure. Previous approaches only addressed 3D anisotropic information indirectly and were limited to two-dimensional, qualitative, strict sample conditions or assumptions.
The research team developed a method enabling the tomographic reconstruction of 3D dielectric tensors without any preparation or assumptions. A sample is illuminated with a laser beam with various angles and circularly polarization states. Then, the light fields scattered from a sample are holographically measured and converted into vectorial diffraction components. Finally, by inversely solving a vectorial wave equation, the 3D dielectric tensor is reconstructed.
Professor YongKeun Park said, “There were a greater number of unknowns in direct measuring than with the conventional approach. We applied our approach to measure additional holographic images by slightly tilting the incident angle.”
He said that the slightly tilted illumination provides an additional orthogonal polarization, which makes the underdetermined problem become the determined problem. “Although scattered fields are dependent on the illumination angle, the Fourier differentiation theorem enables the extraction of the same dielectric tensor for the slightly tilted illumination,” Professor Park added.
His team’s method was validated by reconstructing well-known liquid crystal (LC) structures, including the twisted nematic, hybrid aligned nematic, radial, and bipolar configurations. Furthermore, the research team demonstrated the experimental measurements of the non-equilibrium dynamics of annihilating, nucleating, and merging LC droplets, and the LC polymer network with repeating 3D topological defects.
“This is the first experimental measurement of non-equilibrium dynamics and 3D topological defects in LC structures in a label-free manner. Our method enables the exploration of inaccessible nematic structures and interactions in non-equilibrium dynamics,” first author Dr. Seungwoo Shin explained.
-PublicationSeungwoo Shin, Jonghee Eun, Sang Seok Lee, Changjae Lee, Herve Hugonnet, Dong Ki Yoon, Shin-Hyun Kim, Jongwoo Jeong, YongKeun Park, “Tomographic Measurement ofDielectric Tensors at Optical Frequency,” Nature Materials March 02, 2022 (https://doi.org/10/1038/s41563-022-01202-8)
-ProfileProfessor YongKeun ParkBiomedical Optics Laboratory (http://bmol.kaist.ac.kr)Department of PhysicsCollege of Natural SciencesKAIST
2022.03.22 View 4800
Tomographic Measurement of Dielectric Tensors
Dielectric tensor tomography allows the direct measurement of the 3D dielectric tensors of optically anisotropic structures
A research team reported the direct measurement of dielectric tensors of anisotropic structures including the spatial variations of principal refractive indices and directors. The group also demonstrated quantitative tomographic measurements of various nematic liquid-crystal structures and their fast 3D nonequilibrium dynamics using a 3D label-free tomographic method. The method was described in Nature Materials.
Light-matter interactions are described by the dielectric tensor. Despite their importance in basic science and applications, it has not been possible to measure 3D dielectric tensors directly. The main challenge was due to the vectorial nature of light scattering from a 3D anisotropic structure. Previous approaches only addressed 3D anisotropic information indirectly and were limited to two-dimensional, qualitative, strict sample conditions or assumptions.
The research team developed a method enabling the tomographic reconstruction of 3D dielectric tensors without any preparation or assumptions. A sample is illuminated with a laser beam with various angles and circularly polarization states. Then, the light fields scattered from a sample are holographically measured and converted into vectorial diffraction components. Finally, by inversely solving a vectorial wave equation, the 3D dielectric tensor is reconstructed.
Professor YongKeun Park said, “There were a greater number of unknowns in direct measuring than with the conventional approach. We applied our approach to measure additional holographic images by slightly tilting the incident angle.”
He said that the slightly tilted illumination provides an additional orthogonal polarization, which makes the underdetermined problem become the determined problem. “Although scattered fields are dependent on the illumination angle, the Fourier differentiation theorem enables the extraction of the same dielectric tensor for the slightly tilted illumination,” Professor Park added.
His team’s method was validated by reconstructing well-known liquid crystal (LC) structures, including the twisted nematic, hybrid aligned nematic, radial, and bipolar configurations. Furthermore, the research team demonstrated the experimental measurements of the non-equilibrium dynamics of annihilating, nucleating, and merging LC droplets, and the LC polymer network with repeating 3D topological defects.
“This is the first experimental measurement of non-equilibrium dynamics and 3D topological defects in LC structures in a label-free manner. Our method enables the exploration of inaccessible nematic structures and interactions in non-equilibrium dynamics,” first author Dr. Seungwoo Shin explained.
-PublicationSeungwoo Shin, Jonghee Eun, Sang Seok Lee, Changjae Lee, Herve Hugonnet, Dong Ki Yoon, Shin-Hyun Kim, Jongwoo Jeong, YongKeun Park, “Tomographic Measurement ofDielectric Tensors at Optical Frequency,” Nature Materials March 02, 2022 (https://doi.org/10/1038/s41563-022-01202-8)
-ProfileProfessor YongKeun ParkBiomedical Optics Laboratory (http://bmol.kaist.ac.kr)Department of PhysicsCollege of Natural SciencesKAIST
2022.03.22 View 4800 -
 Scientist Discover How Circadian Rhythm Can Be Both Strong and Flexible
Study reveals that master and slave oscillators function via different molecular mechanisms
From tiny fruit flies to human beings, all animals on Earth maintain their daily rhythms based on their internal circadian clock. The circadian clock enables organisms to undergo rhythmic changes in behavior and physiology based on a 24-hour circadian cycle. For example, our own biological clock tells our brain to release melatonin, a sleep-inducing hormone, at night time.
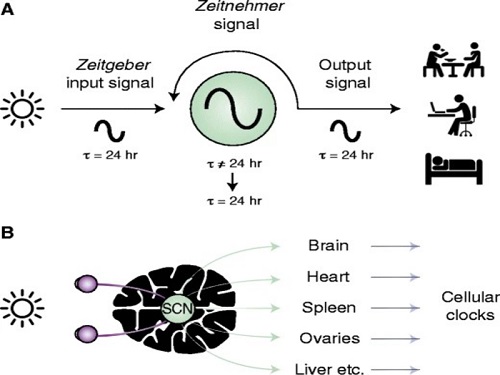
The discovery of the molecular mechanism of the circadian clock was bestowed the Nobel Prize in Physiology or Medicine 2017. From what we know, no one centralized clock is responsible for our circadian cycles. Instead, it operates in a hierarchical network where there are “master pacemaker” and “slave oscillator”.
The master pacemaker receives various input signals from the environment such as light. The master then drives the slave oscillator that regulates various outputs such as sleep, feeding, and metabolism. Despite the different roles of the pacemaker neurons, they are known to share common molecular mechanisms that are well conserved in all lifeforms. For example, interlocked systems of multiple transcriptional-translational feedback loops (TTFLs) composed of core clock proteins have been deeply studied in fruit flies.
However, there is still much that we need to learn about our own biological clock. The hierarchically-organized nature of master and slave clock neurons leads to a prevailing belief that they share an identical molecular clockwork. At the same time, the different roles they serve in regulating bodily rhythms also raise the question of whether they might function under different molecular clockworks.
Research team led by Professor Kim Jae Kyoung from the Department of Mathematical Sciences, a chief investigator at the Biomedical Mathematics Group at the Institute for Basic Science, used a combination of mathematical and experimental approaches using fruit flies to answer this question. The team found that the master clock and the slave clock operate via different molecular mechanisms.
In both master and slave neurons of fruit flies, a circadian rhythm-related protein called PER is produced and degraded at different rates depending on the time of the day. Previously, the team found that the master clock neuron (sLNvs) and the slave clock neuron (DN1ps) have different profiles of PER in wild-type and Clk-Δ mutant Drosophila. This hinted that there might be a potential difference in molecular clockworks between the master and slave clock neurons.
However, due to the complexity of the molecular clockwork, it was challenging to identify the source of such differences. Thus, the team developed a mathematical model describing the molecular clockworks of the master and slave clocks. Then, all possible molecular differences between the master and slave clock neurons were systematically investigated by using computer simulations. The model predicted that PER is more efficiently produced and then rapidly degraded in the master clock compared to the slave clock neurons. This prediction was then confirmed by the follow-up experiments using animal.
Then, why do the master clock neurons have such different molecular properties from the slave clock neurons? To answer this question, the research team again used the combination of mathematical model simulation and experiments. It was found that the faster rate of synthesis of PER in the master clock neurons allows them to generate synchronized rhythms with a high level of amplitude. Generation of such a strong rhythm with high amplitude is critical to delivering clear signals to slave clock neurons.
However, such strong rhythms would typically be unfavorable when it comes to adapting to environmental changes. These include natural causes such as different daylight hours across summer and winter seasons, up to more extreme artificial cases such as jet lag that occurs after international travel. Thanks to the distinct property of the master clock neurons, it is able to undergo phase dispersion when the standard light-dark cycle is disrupted, drastically reducing the level of PER. The master clock neurons can then easily adapt to the new diurnal cycle. Our master pacemaker’s plasticity explains how we can quickly adjust to the new time zones after international flights after just a brief period of jet lag.
It is hoped that the findings of this study can have future clinical implications when it comes to treating various disorders that affect our circadian rhythm. Professor Kim notes, “When the circadian clock loses its robustness and flexibility, the circadian rhythms sleep disorders can occur. As this study identifies the molecular mechanism that generates robustness and flexibility of the circadian clock, it can facilitate the identification of the cause of and treatment strategy for the circadian rhythm sleep disorders.” This work was supported by the Human Frontier Science Program.
-PublicationEui Min Jeong, Miri Kwon, Eunjoo Cho, Sang Hyuk Lee, Hyun Kim, Eun Young Kim, and Jae Kyoung Kim, “Systematic modeling-driven experiments identify distinct molecularclockworks underlying hierarchically organized pacemaker neurons,” February 22, 2022, Proceedings of the National Academy of Sciences of the United States of America
-ProfileProfessor Jae Kyoung KimDepartment of Mathematical SciencesKAIST
2022.02.23 View 5499
Scientist Discover How Circadian Rhythm Can Be Both Strong and Flexible
Study reveals that master and slave oscillators function via different molecular mechanisms
From tiny fruit flies to human beings, all animals on Earth maintain their daily rhythms based on their internal circadian clock. The circadian clock enables organisms to undergo rhythmic changes in behavior and physiology based on a 24-hour circadian cycle. For example, our own biological clock tells our brain to release melatonin, a sleep-inducing hormone, at night time.
The discovery of the molecular mechanism of the circadian clock was bestowed the Nobel Prize in Physiology or Medicine 2017. From what we know, no one centralized clock is responsible for our circadian cycles. Instead, it operates in a hierarchical network where there are “master pacemaker” and “slave oscillator”.
The master pacemaker receives various input signals from the environment such as light. The master then drives the slave oscillator that regulates various outputs such as sleep, feeding, and metabolism. Despite the different roles of the pacemaker neurons, they are known to share common molecular mechanisms that are well conserved in all lifeforms. For example, interlocked systems of multiple transcriptional-translational feedback loops (TTFLs) composed of core clock proteins have been deeply studied in fruit flies.
However, there is still much that we need to learn about our own biological clock. The hierarchically-organized nature of master and slave clock neurons leads to a prevailing belief that they share an identical molecular clockwork. At the same time, the different roles they serve in regulating bodily rhythms also raise the question of whether they might function under different molecular clockworks.
Research team led by Professor Kim Jae Kyoung from the Department of Mathematical Sciences, a chief investigator at the Biomedical Mathematics Group at the Institute for Basic Science, used a combination of mathematical and experimental approaches using fruit flies to answer this question. The team found that the master clock and the slave clock operate via different molecular mechanisms.
In both master and slave neurons of fruit flies, a circadian rhythm-related protein called PER is produced and degraded at different rates depending on the time of the day. Previously, the team found that the master clock neuron (sLNvs) and the slave clock neuron (DN1ps) have different profiles of PER in wild-type and Clk-Δ mutant Drosophila. This hinted that there might be a potential difference in molecular clockworks between the master and slave clock neurons.
However, due to the complexity of the molecular clockwork, it was challenging to identify the source of such differences. Thus, the team developed a mathematical model describing the molecular clockworks of the master and slave clocks. Then, all possible molecular differences between the master and slave clock neurons were systematically investigated by using computer simulations. The model predicted that PER is more efficiently produced and then rapidly degraded in the master clock compared to the slave clock neurons. This prediction was then confirmed by the follow-up experiments using animal.
Then, why do the master clock neurons have such different molecular properties from the slave clock neurons? To answer this question, the research team again used the combination of mathematical model simulation and experiments. It was found that the faster rate of synthesis of PER in the master clock neurons allows them to generate synchronized rhythms with a high level of amplitude. Generation of such a strong rhythm with high amplitude is critical to delivering clear signals to slave clock neurons.
However, such strong rhythms would typically be unfavorable when it comes to adapting to environmental changes. These include natural causes such as different daylight hours across summer and winter seasons, up to more extreme artificial cases such as jet lag that occurs after international travel. Thanks to the distinct property of the master clock neurons, it is able to undergo phase dispersion when the standard light-dark cycle is disrupted, drastically reducing the level of PER. The master clock neurons can then easily adapt to the new diurnal cycle. Our master pacemaker’s plasticity explains how we can quickly adjust to the new time zones after international flights after just a brief period of jet lag.
It is hoped that the findings of this study can have future clinical implications when it comes to treating various disorders that affect our circadian rhythm. Professor Kim notes, “When the circadian clock loses its robustness and flexibility, the circadian rhythms sleep disorders can occur. As this study identifies the molecular mechanism that generates robustness and flexibility of the circadian clock, it can facilitate the identification of the cause of and treatment strategy for the circadian rhythm sleep disorders.” This work was supported by the Human Frontier Science Program.
-PublicationEui Min Jeong, Miri Kwon, Eunjoo Cho, Sang Hyuk Lee, Hyun Kim, Eun Young Kim, and Jae Kyoung Kim, “Systematic modeling-driven experiments identify distinct molecularclockworks underlying hierarchically organized pacemaker neurons,” February 22, 2022, Proceedings of the National Academy of Sciences of the United States of America
-ProfileProfessor Jae Kyoung KimDepartment of Mathematical SciencesKAIST
2022.02.23 View 5499 -
 Label-Free Multiplexed Microtomography of Endogenous Subcellular Dynamics Using Deep Learning
AI-based holographic microscopy allows molecular imaging without introducing exogenous labeling agents
A research team upgraded the 3D microtomography observing dynamics of label-free live cells in multiplexed fluorescence imaging. The AI-powered 3D holotomographic microscopy extracts various molecular information from live unlabeled biological cells in real time without exogenous labeling or staining agents.
Professor YongKeum Park’s team and the startup Tomocube encoded 3D refractive index tomograms using the refractive index as a means of measurement. Then they decoded the information with a deep learning-based model that infers multiple 3D fluorescence tomograms from the refractive index measurements of the corresponding subcellular targets, thereby achieving multiplexed micro tomography. This study was reported in Nature Cell Biology online on December 7, 2021.
Fluorescence microscopy is the most widely used optical microscopy technique due to its high biochemical specificity. However, it needs to genetically manipulate or to stain cells with fluorescent labels in order to express fluorescent proteins. These labeling processes inevitably affect the intrinsic physiology of cells. It also has challenges in long-term measuring due to photobleaching and phototoxicity. The overlapped spectra of multiplexed fluorescence signals also hinder the viewing of various structures at the same time. More critically, it took several hours to observe the cells after preparing them.
3D holographic microscopy, also known as holotomography, is providing new ways to quantitatively image live cells without pretreatments such as staining. Holotomography can accurately and quickly measure the morphological and structural information of cells, but only provides limited biochemical and molecular information.
The 'AI microscope' created in this process takes advantage of the features of both holographic microscopy and fluorescence microscopy. That is, a specific image from a fluorescence microscope can be obtained without a fluorescent label. Therefore, the microscope can observe many types of cellular structures in their natural state in 3D and at the same time as fast as one millisecond, and long-term measurements over several days are also possible.
The Tomocube-KAIST team showed that fluorescence images can be directly and precisely predicted from holotomographic images in various cells and conditions. Using the quantitative relationship between the spatial distribution of the refractive index found by AI and the major structures in cells, it was possible to decipher the spatial distribution of the refractive index. And surprisingly, it confirmed that this relationship is constant regardless of cell type.
Professor Park said, “We were able to develop a new concept microscope that combines the advantages of several microscopes with the multidisciplinary research of AI, optics, and biology. It will be immediately applicable for new types of cells not included in the existing data and is expected to be widely applicable for various biological and medical research.”
When comparing the molecular image information extracted by AI with the molecular image information physically obtained by fluorescence staining in 3D space, it showed a 97% or more conformity, which is a level that is difficult to distinguish with the naked eye.
“Compared to the sub-60% accuracy of the fluorescence information extracted from the model developed by the Google AI team, it showed significantly higher performance,” Professor Park added.
This work was supported by the KAIST Up program, the BK21+ program, Tomocube, the National Research Foundation of Korea, and the Ministry of Science and ICT, and the Ministry of Health & Welfare.
-Publication
Hyun-seok Min, Won-Do Heo, YongKeun Park, et al. “Label-free multiplexed microtomography of endogenous subcellular dynamics using generalizable deep learning,” Nature Cell Biology (doi.org/10.1038/s41556-021-00802-x) published online December 07 2021.
-Profile
Professor YongKeun Park
Biomedical Optics Laboratory
Department of Physics
KAIST
2022.02.09 View 6341
Label-Free Multiplexed Microtomography of Endogenous Subcellular Dynamics Using Deep Learning
AI-based holographic microscopy allows molecular imaging without introducing exogenous labeling agents
A research team upgraded the 3D microtomography observing dynamics of label-free live cells in multiplexed fluorescence imaging. The AI-powered 3D holotomographic microscopy extracts various molecular information from live unlabeled biological cells in real time without exogenous labeling or staining agents.
Professor YongKeum Park’s team and the startup Tomocube encoded 3D refractive index tomograms using the refractive index as a means of measurement. Then they decoded the information with a deep learning-based model that infers multiple 3D fluorescence tomograms from the refractive index measurements of the corresponding subcellular targets, thereby achieving multiplexed micro tomography. This study was reported in Nature Cell Biology online on December 7, 2021.
Fluorescence microscopy is the most widely used optical microscopy technique due to its high biochemical specificity. However, it needs to genetically manipulate or to stain cells with fluorescent labels in order to express fluorescent proteins. These labeling processes inevitably affect the intrinsic physiology of cells. It also has challenges in long-term measuring due to photobleaching and phototoxicity. The overlapped spectra of multiplexed fluorescence signals also hinder the viewing of various structures at the same time. More critically, it took several hours to observe the cells after preparing them.
3D holographic microscopy, also known as holotomography, is providing new ways to quantitatively image live cells without pretreatments such as staining. Holotomography can accurately and quickly measure the morphological and structural information of cells, but only provides limited biochemical and molecular information.
The 'AI microscope' created in this process takes advantage of the features of both holographic microscopy and fluorescence microscopy. That is, a specific image from a fluorescence microscope can be obtained without a fluorescent label. Therefore, the microscope can observe many types of cellular structures in their natural state in 3D and at the same time as fast as one millisecond, and long-term measurements over several days are also possible.
The Tomocube-KAIST team showed that fluorescence images can be directly and precisely predicted from holotomographic images in various cells and conditions. Using the quantitative relationship between the spatial distribution of the refractive index found by AI and the major structures in cells, it was possible to decipher the spatial distribution of the refractive index. And surprisingly, it confirmed that this relationship is constant regardless of cell type.
Professor Park said, “We were able to develop a new concept microscope that combines the advantages of several microscopes with the multidisciplinary research of AI, optics, and biology. It will be immediately applicable for new types of cells not included in the existing data and is expected to be widely applicable for various biological and medical research.”
When comparing the molecular image information extracted by AI with the molecular image information physically obtained by fluorescence staining in 3D space, it showed a 97% or more conformity, which is a level that is difficult to distinguish with the naked eye.
“Compared to the sub-60% accuracy of the fluorescence information extracted from the model developed by the Google AI team, it showed significantly higher performance,” Professor Park added.
This work was supported by the KAIST Up program, the BK21+ program, Tomocube, the National Research Foundation of Korea, and the Ministry of Science and ICT, and the Ministry of Health & Welfare.
-Publication
Hyun-seok Min, Won-Do Heo, YongKeun Park, et al. “Label-free multiplexed microtomography of endogenous subcellular dynamics using generalizable deep learning,” Nature Cell Biology (doi.org/10.1038/s41556-021-00802-x) published online December 07 2021.
-Profile
Professor YongKeun Park
Biomedical Optics Laboratory
Department of Physics
KAIST
2022.02.09 View 6341 -
 Connecting the Dots to Find New Treatments for Breast Cancer
Systems biologists uncovered new ways of cancer cell reprogramming to treat drug-resistant cancers
Scientists at KAIST believe they may have found a way to reverse an aggressive, treatment-resistant type of breast cancer into a less dangerous kind that responds well to treatment. The study involved the use of mathematical models to untangle the complex genetic and molecular interactions that occur in the two types of breast cancer, but could be extended to find ways for treating many others. The study’s findings were published in the journal Cancer Research.
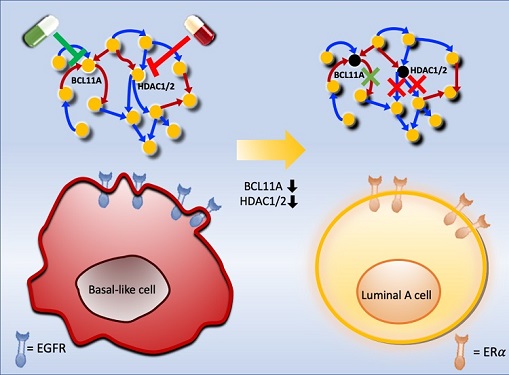
Basal-like tumours are the most aggressive type of breast cancer, with the worst prognosis. Chemotherapy is the only available treatment option, but patients experience high recurrence rates. On the other hand, luminal-A breast cancer responds well to drugs that specifically target a receptor on their cell surfaces, called estrogen receptor alpha (ERα).
KAIST systems biologist Kwang-Hyun Cho and colleagues analyzed the complex molecular and genetic interactions of basal-like and luminal-A breast cancers to find out if there might be a way to switch the former to the latter and give patients a better chance to respond to treatment.
To do this, they accessed large amounts of cancer and patient data to understand which genes and molecules are involved in the two types. They then input this data into a mathematical model that represents genes, proteins and molecules as dots and the interactions between them as lines. The model can be used to conduct simulations and see how interactions change when certain genes are turned on or off.
“There have been a tremendous number of studies trying to find therapeutic targets for treating basal-like breast cancer patients,” says Cho. “But clinical trials have failed due to the complex and dynamic nature of cancer. To overcome this issue, we looked at breast cancer cells as a complex network system and implemented a systems biological approach to unravel the underlying mechanisms that would allow us to reprogram basal-like into luminal-A breast cancer cells.”
Using this approach, followed by experimental validation on real breast cancer cells, the team found that turning off two key gene regulators, called BCL11A and HDAC1/2, switched a basal-like cancer signalling pathway into a different one used by luminal-A cancer cells. The switch reprograms the cancer cells and makes them more responsive to drugs that target ERα receptors. However, further tests will be needed to confirm that this also works in animal models and eventually humans.
“Our study demonstrates that the systems biological approach can be useful for identifying novel therapeutic targets,” says Cho.
The researchers are now expanding its breast cancer network model to include all breast cancer subtypes. Their ultimate aim is to identify more drug targets and to understand the mechanisms that could drive drug-resistant cells to turn into drug-sensitive ones.
This work was supported by the National Research Foundation of Korea, the Ministry of Science and ICT, Electronics and Telecommunications Research Institute, and the KAIST Grand Challenge 30 Project.
-Publication Sea R. Choi, Chae Young Hwang, Jonghoon Lee, and Kwang-Hyun Cho, “Network Analysis Identifies Regulators of Basal-like Breast Cancer Reprogramming and Endocrine TherapyVulnerability,” Cancer Research, November 30. (doi:10.1158/0008-5472.CAN-21-0621)
-ProfileProfessor Kwang-Hyun ChoLaboratory for Systems Biology and Bio-Inspired EngineeringDepartment of Bio and Brain EngineeringKAIST
2021.12.07 View 5919
Connecting the Dots to Find New Treatments for Breast Cancer
Systems biologists uncovered new ways of cancer cell reprogramming to treat drug-resistant cancers
Scientists at KAIST believe they may have found a way to reverse an aggressive, treatment-resistant type of breast cancer into a less dangerous kind that responds well to treatment. The study involved the use of mathematical models to untangle the complex genetic and molecular interactions that occur in the two types of breast cancer, but could be extended to find ways for treating many others. The study’s findings were published in the journal Cancer Research.
Basal-like tumours are the most aggressive type of breast cancer, with the worst prognosis. Chemotherapy is the only available treatment option, but patients experience high recurrence rates. On the other hand, luminal-A breast cancer responds well to drugs that specifically target a receptor on their cell surfaces, called estrogen receptor alpha (ERα).
KAIST systems biologist Kwang-Hyun Cho and colleagues analyzed the complex molecular and genetic interactions of basal-like and luminal-A breast cancers to find out if there might be a way to switch the former to the latter and give patients a better chance to respond to treatment.
To do this, they accessed large amounts of cancer and patient data to understand which genes and molecules are involved in the two types. They then input this data into a mathematical model that represents genes, proteins and molecules as dots and the interactions between them as lines. The model can be used to conduct simulations and see how interactions change when certain genes are turned on or off.
“There have been a tremendous number of studies trying to find therapeutic targets for treating basal-like breast cancer patients,” says Cho. “But clinical trials have failed due to the complex and dynamic nature of cancer. To overcome this issue, we looked at breast cancer cells as a complex network system and implemented a systems biological approach to unravel the underlying mechanisms that would allow us to reprogram basal-like into luminal-A breast cancer cells.”
Using this approach, followed by experimental validation on real breast cancer cells, the team found that turning off two key gene regulators, called BCL11A and HDAC1/2, switched a basal-like cancer signalling pathway into a different one used by luminal-A cancer cells. The switch reprograms the cancer cells and makes them more responsive to drugs that target ERα receptors. However, further tests will be needed to confirm that this also works in animal models and eventually humans.
“Our study demonstrates that the systems biological approach can be useful for identifying novel therapeutic targets,” says Cho.
The researchers are now expanding its breast cancer network model to include all breast cancer subtypes. Their ultimate aim is to identify more drug targets and to understand the mechanisms that could drive drug-resistant cells to turn into drug-sensitive ones.
This work was supported by the National Research Foundation of Korea, the Ministry of Science and ICT, Electronics and Telecommunications Research Institute, and the KAIST Grand Challenge 30 Project.
-Publication Sea R. Choi, Chae Young Hwang, Jonghoon Lee, and Kwang-Hyun Cho, “Network Analysis Identifies Regulators of Basal-like Breast Cancer Reprogramming and Endocrine TherapyVulnerability,” Cancer Research, November 30. (doi:10.1158/0008-5472.CAN-21-0621)
-ProfileProfessor Kwang-Hyun ChoLaboratory for Systems Biology and Bio-Inspired EngineeringDepartment of Bio and Brain EngineeringKAIST
2021.12.07 View 5919 -
 Scientists Develop Wireless Networks that Allow Brain Circuits to Be Controlled Remotely through the Internet
Wireless implantable devices and IoT could manipulate the brains of animals from anywhere around the world due to their minimalistic hardware, low setup cost, ease of use, and customizable versatility
A new study shows that researchers can remotely control the brain circuits of numerous animals simultaneously and independently through the internet. The scientists believe this newly developed technology can speed up brain research and various neuroscience studies to uncover basic brain functions as well as the underpinnings of various neuropsychiatric and neurological disorders.
A multidisciplinary team of researchers at KAIST, Washington University in St. Louis, and the University of Colorado, Boulder, created a wireless ecosystem with its own wireless implantable devices and Internet of Things (IoT) infrastructure to enable high-throughput neuroscience experiments over the internet. This innovative technology could enable scientists to manipulate the brains of animals from anywhere around the world. The study was published in the journal Nature Biomedical Engineering on November 25
“This novel technology is highly versatile and adaptive. It can remotely control numerous neural implants and laboratory tools in real-time or in a scheduled way without direct human interactions,” said Professor Jae-Woong Jeong of the School of Electrical Engineering at KAIST and a senior author of the study. “These wireless neural devices and equipment integrated with IoT technology have enormous potential for science and medicine.”
The wireless ecosystem only requires a mini-computer that can be purchased for under $45, which connects to the internet and communicates with wireless multifunctional brain probes or other types of conventional laboratory equipment using IoT control modules. By optimally integrating the versatility and modular construction of both unique IoT hardware and software within a single ecosystem, this wireless technology offers new applications that have not been demonstrated before by a single standalone technology. This includes, but is not limited to minimalistic hardware, global remote access, selective and scheduled experiments, customizable automation, and high-throughput scalability.
“As long as researchers have internet access, they are able to trigger, customize, stop, validate, and store the outcomes of large experiments at any time and from anywhere in the world. They can remotely perform large-scale neuroscience experiments in animals deployed in multiple countries,” said one of the lead authors, Dr. Raza Qazi, a researcher with KAIST and the University of Colorado, Boulder. “The low cost of this system allows it to be easily adopted and can further fuel innovation across many laboratories,” Dr. Qazi added.
One of the significant advantages of this IoT neurotechnology is its ability to be mass deployed across the globe due to its minimalistic hardware, low setup cost, ease of use, and customizable versatility. Scientists across the world can quickly implement this technology within their existing laboratories with minimal budget concerns to achieve globally remote access, scalable experimental automation, or both, thus potentially reducing the time needed to unravel various neuroscientific challenges such as those associated with intractable neurological conditions.
Another senior author on the study, Professor Jordan McCall from the Department of Anesthesiology and Center for Clinical Pharmacology at Washington University in St. Louis, said this technology has the potential to change how basic neuroscience studies are performed. “One of the biggest limitations when trying to understand how the mammalian brain works is that we have to study these functions in unnatural conditions. This technology brings us one step closer to performing important studies without direct human interaction with the study subjects.”
The ability to remotely schedule experiments moves toward automating these types of experiments. Dr. Kyle Parker, an instructor at Washington University in St. Louis and another lead author on the study added, “This experimental automation can potentially help us reduce the number of animals used in biomedical research by reducing the variability introduced by various experimenters. This is especially important given our moral imperative to seek research designs that enable this reduction.”
The researchers believe this wireless technology may open new opportunities for many applications including brain research, pharmaceuticals, and telemedicine to treat diseases in the brain and other organs remotely. This remote automation technology could become even more valuable when many labs need to shut down, such as during the height of the COVID-19 pandemic.
This work was supported by grants from the KAIST Global Singularity Research Program, the National Research Foundation of Korea, the United States National Institute of Health, and Oak Ridge Associated Universities.
-PublicationRaza Qazi, Kyle Parker, Choong Yeon Kim, Jordan McCall, Jae-Woong Jeong et al. “Scalable and modular wireless-network infrastructure for large-scale behavioral neuroscience,” Nature Biomedical Engineering, November 25 2021 (doi.org/10.1038/s41551-021-00814-w)
-ProfileProfessor Jae-Woong JeongBio-Integrated Electronics and Systems LabSchool of Electrical EngineeringKAIST
2021.11.29 View 9849
Scientists Develop Wireless Networks that Allow Brain Circuits to Be Controlled Remotely through the Internet
Wireless implantable devices and IoT could manipulate the brains of animals from anywhere around the world due to their minimalistic hardware, low setup cost, ease of use, and customizable versatility
A new study shows that researchers can remotely control the brain circuits of numerous animals simultaneously and independently through the internet. The scientists believe this newly developed technology can speed up brain research and various neuroscience studies to uncover basic brain functions as well as the underpinnings of various neuropsychiatric and neurological disorders.
A multidisciplinary team of researchers at KAIST, Washington University in St. Louis, and the University of Colorado, Boulder, created a wireless ecosystem with its own wireless implantable devices and Internet of Things (IoT) infrastructure to enable high-throughput neuroscience experiments over the internet. This innovative technology could enable scientists to manipulate the brains of animals from anywhere around the world. The study was published in the journal Nature Biomedical Engineering on November 25
“This novel technology is highly versatile and adaptive. It can remotely control numerous neural implants and laboratory tools in real-time or in a scheduled way without direct human interactions,” said Professor Jae-Woong Jeong of the School of Electrical Engineering at KAIST and a senior author of the study. “These wireless neural devices and equipment integrated with IoT technology have enormous potential for science and medicine.”
The wireless ecosystem only requires a mini-computer that can be purchased for under $45, which connects to the internet and communicates with wireless multifunctional brain probes or other types of conventional laboratory equipment using IoT control modules. By optimally integrating the versatility and modular construction of both unique IoT hardware and software within a single ecosystem, this wireless technology offers new applications that have not been demonstrated before by a single standalone technology. This includes, but is not limited to minimalistic hardware, global remote access, selective and scheduled experiments, customizable automation, and high-throughput scalability.
“As long as researchers have internet access, they are able to trigger, customize, stop, validate, and store the outcomes of large experiments at any time and from anywhere in the world. They can remotely perform large-scale neuroscience experiments in animals deployed in multiple countries,” said one of the lead authors, Dr. Raza Qazi, a researcher with KAIST and the University of Colorado, Boulder. “The low cost of this system allows it to be easily adopted and can further fuel innovation across many laboratories,” Dr. Qazi added.
One of the significant advantages of this IoT neurotechnology is its ability to be mass deployed across the globe due to its minimalistic hardware, low setup cost, ease of use, and customizable versatility. Scientists across the world can quickly implement this technology within their existing laboratories with minimal budget concerns to achieve globally remote access, scalable experimental automation, or both, thus potentially reducing the time needed to unravel various neuroscientific challenges such as those associated with intractable neurological conditions.
Another senior author on the study, Professor Jordan McCall from the Department of Anesthesiology and Center for Clinical Pharmacology at Washington University in St. Louis, said this technology has the potential to change how basic neuroscience studies are performed. “One of the biggest limitations when trying to understand how the mammalian brain works is that we have to study these functions in unnatural conditions. This technology brings us one step closer to performing important studies without direct human interaction with the study subjects.”
The ability to remotely schedule experiments moves toward automating these types of experiments. Dr. Kyle Parker, an instructor at Washington University in St. Louis and another lead author on the study added, “This experimental automation can potentially help us reduce the number of animals used in biomedical research by reducing the variability introduced by various experimenters. This is especially important given our moral imperative to seek research designs that enable this reduction.”
The researchers believe this wireless technology may open new opportunities for many applications including brain research, pharmaceuticals, and telemedicine to treat diseases in the brain and other organs remotely. This remote automation technology could become even more valuable when many labs need to shut down, such as during the height of the COVID-19 pandemic.
This work was supported by grants from the KAIST Global Singularity Research Program, the National Research Foundation of Korea, the United States National Institute of Health, and Oak Ridge Associated Universities.
-PublicationRaza Qazi, Kyle Parker, Choong Yeon Kim, Jordan McCall, Jae-Woong Jeong et al. “Scalable and modular wireless-network infrastructure for large-scale behavioral neuroscience,” Nature Biomedical Engineering, November 25 2021 (doi.org/10.1038/s41551-021-00814-w)
-ProfileProfessor Jae-Woong JeongBio-Integrated Electronics and Systems LabSchool of Electrical EngineeringKAIST
2021.11.29 View 9849 -
 The Dynamic Tracking of Tissue-Specific Secretory Proteins
Researchers develop a versatile and powerful tool for studying the spatiotemporal dynamics of secretory proteins, a valuable class of biomarkers and therapeutic targets
Researchers have presented a method for profiling tissue-specific secretory proteins in live mice. This method is expected to be applicable to various tissues or disease models for investigating biomarkers or therapeutic targets involved in disease progression. This research was reported in Nature Communications on September 1.
Secretory proteins released into the blood play essential roles in physiological systems. They are core mediators of interorgan communication, while serving as biomarkers and therapeutic targets.
Previous studies have analyzed conditioned media from culture models to identify cell type-specific secretory proteins, but these models often fail to fully recapitulate the intricacies of multi-organ systems and thus do not sufficiently reflect biological realities.
These limitations provided compelling motivation for the research team led by Jae Myoung Suh and his collaborators to develop techniques that could identify and resolve characteristics of tissue-specific secretory proteins along time and space dimensions.
For addressing this gap in the current methodology, the research team utilized proximity-labeling enzymes such as TurboID to label secretory proteins in endoplasmic reticulum lumen using biotin. Thereafter, the biotin-labeled secretory proteins were readily enriched through streptavidin affinity purification and could be identified through mass spectrometry.
To demonstrate its functionality in live mice, research team delivered TurboID to mouse livers via an adenovirus. After administering the biotin, only liver-derived secretory proteins were successfully detected in the plasma of the mice. Interestingly, the pattern of biotin-labeled proteins secreted from the liver was clearly distinctive from those of hepatocyte cell lines.
First author Kwang-eun Kim from the Graduate School of Medical Science and Engineering explained, “The proteins secreted by the liver were significantly different from the results of cell culture models. This data shows the limitations of cell culture models for secretory protein study, and this technique can overcome those limitations. It can be further used to discover biomarkers and therapeutic targets that can more fully reflect the physiological state.”
This work research was supported by the National Research Foundation of Korea, the KAIST Key Research Institutes Project (Interdisciplinary Research Group), and the Institute for Basic Science in Korea.
-PublicationKwang-eun Kim, Isaac Park et al., “Dynamic tracking and identification of tissue-specific secretory proteins in the circulation of live mice,” Nature Communications on Sept.1,
2021(https://doi.org/10.1038/s41467-021-25546-y)
-ProfileProfessor Jae Myoung Suh Integrated Lab of Metabolism, Obesity and Diabetes Researchhttps://imodkaist.wixsite.com/home
Graduate School of Medical Science and Engineering College of Life Science and BioengineeringKAIST
2021.09.14 View 6005
The Dynamic Tracking of Tissue-Specific Secretory Proteins
Researchers develop a versatile and powerful tool for studying the spatiotemporal dynamics of secretory proteins, a valuable class of biomarkers and therapeutic targets
Researchers have presented a method for profiling tissue-specific secretory proteins in live mice. This method is expected to be applicable to various tissues or disease models for investigating biomarkers or therapeutic targets involved in disease progression. This research was reported in Nature Communications on September 1.
Secretory proteins released into the blood play essential roles in physiological systems. They are core mediators of interorgan communication, while serving as biomarkers and therapeutic targets.
Previous studies have analyzed conditioned media from culture models to identify cell type-specific secretory proteins, but these models often fail to fully recapitulate the intricacies of multi-organ systems and thus do not sufficiently reflect biological realities.
These limitations provided compelling motivation for the research team led by Jae Myoung Suh and his collaborators to develop techniques that could identify and resolve characteristics of tissue-specific secretory proteins along time and space dimensions.
For addressing this gap in the current methodology, the research team utilized proximity-labeling enzymes such as TurboID to label secretory proteins in endoplasmic reticulum lumen using biotin. Thereafter, the biotin-labeled secretory proteins were readily enriched through streptavidin affinity purification and could be identified through mass spectrometry.
To demonstrate its functionality in live mice, research team delivered TurboID to mouse livers via an adenovirus. After administering the biotin, only liver-derived secretory proteins were successfully detected in the plasma of the mice. Interestingly, the pattern of biotin-labeled proteins secreted from the liver was clearly distinctive from those of hepatocyte cell lines.
First author Kwang-eun Kim from the Graduate School of Medical Science and Engineering explained, “The proteins secreted by the liver were significantly different from the results of cell culture models. This data shows the limitations of cell culture models for secretory protein study, and this technique can overcome those limitations. It can be further used to discover biomarkers and therapeutic targets that can more fully reflect the physiological state.”
This work research was supported by the National Research Foundation of Korea, the KAIST Key Research Institutes Project (Interdisciplinary Research Group), and the Institute for Basic Science in Korea.
-PublicationKwang-eun Kim, Isaac Park et al., “Dynamic tracking and identification of tissue-specific secretory proteins in the circulation of live mice,” Nature Communications on Sept.1,
2021(https://doi.org/10.1038/s41467-021-25546-y)
-ProfileProfessor Jae Myoung Suh Integrated Lab of Metabolism, Obesity and Diabetes Researchhttps://imodkaist.wixsite.com/home
Graduate School of Medical Science and Engineering College of Life Science and BioengineeringKAIST
2021.09.14 View 6005 -
 A Mechanism Underlying Most Common Cause of Epileptic Seizures Revealed
An interdisciplinary study shows that neurons carrying somatic mutations in MTOR can lead to focal epileptogenesis via non-cell-autonomous hyperexcitability of nearby nonmutated neurons
During fetal development, cells should migrate to the outer edge of the brain to form critical connections for information transfer and regulation in the body. When even a few cells fail to move to the correct location, the neurons become disorganized and this results in focal cortical dysplasia. This condition is the most common cause of seizures that cannot be controlled with medication in children and the second most common cause in adults.
Now, an interdisciplinary team studying neurogenetics, neural networks, and neurophysiology at KAIST has revealed how dysfunctions in even a small percentage of cells can cause disorder across the entire brain. They published their results on June 28 in Annals of Neurology.
The work builds on a previous finding, also by a KAIST scientists, who found that focal cortical dysplasia was caused by mutations in the cells involved in mTOR, a pathway that regulates signaling between neurons in the brain.
“Only 1 to 2% of neurons carrying mutations in the mTOR signaling pathway that regulates cell signaling in the brain have been found to include seizures in animal models of focal cortical dysplasia,” said Professor Jong-Woo Sohn from the Department of Biological Sciences. “The main challenge of this study was to explain how nearby non-mutated neurons are hyperexcitable.”
Initially, the researchers hypothesized that the mutated cells affected the number of excitatory and inhibitory synapses in all neurons, mutated or not. These neural gates can trigger or halt activity, respectively, in other neurons. Seizures are a result of extreme activity, called hyperexcitability. If the mutated cells upend the balance and result in more excitatory cells, the researchers thought, it made sense that the cells would be more susceptible to hyperexcitability and, as a result, seizures.
“Contrary to our expectations, the synaptic input balance was not changed in either the mutated or non-mutated neurons,” said Professor Jeong Ho Lee from the Graduate School of Medical Science and Engineering. “We turned our attention to a protein overproduced by mutated neurons.”
The protein is adenosine kinase, which lowers the concentration of adenosine. This naturally occurring compound is an anticonvulsant and works to relax vessels. In mice engineered to have focal cortical dysplasia, the researchers injected adenosine to replace the levels lowered by the protein. It worked and the neurons became less excitable.
“We demonstrated that augmentation of adenosine signaling could attenuate the excitability of non-mutated neurons,” said Professor Se-Bum Paik from the Department of Bio and Brain Engineering.
The effect on the non-mutated neurons was the surprising part, according to Paik. “The seizure-triggering hyperexcitability originated not in the mutation-carrying neurons, but instead in the nearby non-mutated neurons,” he said.
The mutated neurons excreted more adenosine kinase, reducing the adenosine levels in the local environment of all the cells. With less adenosine, the non-mutated neurons became hyperexcitable, leading to seizures.
“While we need further investigate into the relationship between the concentration of adenosine and the increased excitation of nearby neurons, our results support the medical use of drugs to activate adenosine signaling as a possible treatment pathway for focal cortical dysplasia,” Professor Lee said.
The Suh Kyungbae Foundation, the Korea Health Technology Research and Development Project, the Ministry of Health & Welfare, and the National Research Foundation in Korea funded this work.
-Publication:Koh, H.Y., Jang, J., Ju, S.H., Kim, R., Cho, G.-B., Kim, D.S., Sohn, J.-W., Paik, S.-B. and Lee, J.H. (2021), ‘Non–Cell Autonomous Epileptogenesis in Focal Cortical Dysplasia’ Annals of Neurology, 90: 285 299. (https://doi.org/10.1002/ana.26149)
-ProfileProfessor Jeong Ho Lee Translational Neurogenetics Labhttps://tnl.kaist.ac.kr/ Graduate School of Medical Science and Engineering KAIST
Professor Se-Bum Paik Visual System and Neural Network Laboratory http://vs.kaist.ac.kr/ Department of Bio and Brain EngineeringKAIST
Professor Jong-Woo Sohn Laboratory for Neurophysiology, https://sites.google.com/site/sohnlab2014/home Department of Biological SciencesKAIST
Dr. Hyun Yong Koh Translational Neurogenetics LabGraduate School of Medical Science and EngineeringKAIST
Dr. Jaeson Jang Ph.D.Visual System and Neural Network LaboratoryDepartment of Bio and Brain Engineering KAIST
Sang Hyeon Ju M.D.Laboratory for NeurophysiologyDepartment of Biological SciencesKAIST
2021.08.26 View 8352
A Mechanism Underlying Most Common Cause of Epileptic Seizures Revealed
An interdisciplinary study shows that neurons carrying somatic mutations in MTOR can lead to focal epileptogenesis via non-cell-autonomous hyperexcitability of nearby nonmutated neurons
During fetal development, cells should migrate to the outer edge of the brain to form critical connections for information transfer and regulation in the body. When even a few cells fail to move to the correct location, the neurons become disorganized and this results in focal cortical dysplasia. This condition is the most common cause of seizures that cannot be controlled with medication in children and the second most common cause in adults.
Now, an interdisciplinary team studying neurogenetics, neural networks, and neurophysiology at KAIST has revealed how dysfunctions in even a small percentage of cells can cause disorder across the entire brain. They published their results on June 28 in Annals of Neurology.
The work builds on a previous finding, also by a KAIST scientists, who found that focal cortical dysplasia was caused by mutations in the cells involved in mTOR, a pathway that regulates signaling between neurons in the brain.
“Only 1 to 2% of neurons carrying mutations in the mTOR signaling pathway that regulates cell signaling in the brain have been found to include seizures in animal models of focal cortical dysplasia,” said Professor Jong-Woo Sohn from the Department of Biological Sciences. “The main challenge of this study was to explain how nearby non-mutated neurons are hyperexcitable.”
Initially, the researchers hypothesized that the mutated cells affected the number of excitatory and inhibitory synapses in all neurons, mutated or not. These neural gates can trigger or halt activity, respectively, in other neurons. Seizures are a result of extreme activity, called hyperexcitability. If the mutated cells upend the balance and result in more excitatory cells, the researchers thought, it made sense that the cells would be more susceptible to hyperexcitability and, as a result, seizures.
“Contrary to our expectations, the synaptic input balance was not changed in either the mutated or non-mutated neurons,” said Professor Jeong Ho Lee from the Graduate School of Medical Science and Engineering. “We turned our attention to a protein overproduced by mutated neurons.”
The protein is adenosine kinase, which lowers the concentration of adenosine. This naturally occurring compound is an anticonvulsant and works to relax vessels. In mice engineered to have focal cortical dysplasia, the researchers injected adenosine to replace the levels lowered by the protein. It worked and the neurons became less excitable.
“We demonstrated that augmentation of adenosine signaling could attenuate the excitability of non-mutated neurons,” said Professor Se-Bum Paik from the Department of Bio and Brain Engineering.
The effect on the non-mutated neurons was the surprising part, according to Paik. “The seizure-triggering hyperexcitability originated not in the mutation-carrying neurons, but instead in the nearby non-mutated neurons,” he said.
The mutated neurons excreted more adenosine kinase, reducing the adenosine levels in the local environment of all the cells. With less adenosine, the non-mutated neurons became hyperexcitable, leading to seizures.
“While we need further investigate into the relationship between the concentration of adenosine and the increased excitation of nearby neurons, our results support the medical use of drugs to activate adenosine signaling as a possible treatment pathway for focal cortical dysplasia,” Professor Lee said.
The Suh Kyungbae Foundation, the Korea Health Technology Research and Development Project, the Ministry of Health & Welfare, and the National Research Foundation in Korea funded this work.
-Publication:Koh, H.Y., Jang, J., Ju, S.H., Kim, R., Cho, G.-B., Kim, D.S., Sohn, J.-W., Paik, S.-B. and Lee, J.H. (2021), ‘Non–Cell Autonomous Epileptogenesis in Focal Cortical Dysplasia’ Annals of Neurology, 90: 285 299. (https://doi.org/10.1002/ana.26149)
-ProfileProfessor Jeong Ho Lee Translational Neurogenetics Labhttps://tnl.kaist.ac.kr/ Graduate School of Medical Science and Engineering KAIST
Professor Se-Bum Paik Visual System and Neural Network Laboratory http://vs.kaist.ac.kr/ Department of Bio and Brain EngineeringKAIST
Professor Jong-Woo Sohn Laboratory for Neurophysiology, https://sites.google.com/site/sohnlab2014/home Department of Biological SciencesKAIST
Dr. Hyun Yong Koh Translational Neurogenetics LabGraduate School of Medical Science and EngineeringKAIST
Dr. Jaeson Jang Ph.D.Visual System and Neural Network LaboratoryDepartment of Bio and Brain Engineering KAIST
Sang Hyeon Ju M.D.Laboratory for NeurophysiologyDepartment of Biological SciencesKAIST
2021.08.26 View 8352 -
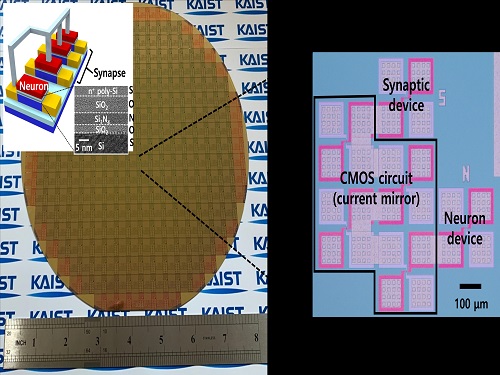 Brain-Inspired Highly Scalable Neuromorphic Hardware Presented
Neurons and synapses based on single transistor can dramatically reduce the hardware cost and accelerate the commercialization of neuromorphic hardware
KAIST researchers fabricated a brain-inspired highly scalable neuromorphic hardware by co-integrating single transistor neurons and synapses. Using standard silicon complementary metal-oxide-semiconductor (CMOS) technology, the neuromorphic hardware is expected to reduce chip cost and simplify fabrication procedures.
The research team led by Yang-Kyu Choi and Sung-Yool Choi produced a neurons and synapses based on single transistor for highly scalable neuromorphic hardware and showed the ability to recognize text and face images. This research was featured in Science Advances on August 4.
Neuromorphic hardware has attracted a great deal of attention because of its artificial intelligence functions, but consuming ultra-low power of less than 20 watts by mimicking the human brain. To make neuromorphic hardware work, a neuron that generates a spike when integrating a certain signal, and a synapse remembering the connection between two neurons are necessary, just like the biological brain. However, since neurons and synapses constructed on digital or analog circuits occupy a large space, there is a limit in terms of hardware efficiency and costs. Since the human brain consists of about 1011 neurons and 1014 synapses, it is necessary to improve the hardware cost in order to apply it to mobile and IoT devices.
To solve the problem, the research team mimicked the behavior of biological neurons and synapses with a single transistor, and co-integrated them onto an 8-inch wafer. The manufactured neuromorphic transistors have the same structure as the transistors for memory and logic that are currently mass-produced. In addition, the neuromorphic transistors proved for the first time that they can be implemented with a ‘Janus structure’ that functions as both neuron and synapse, just like coins have heads and tails.
Professor Yang-Kyu Choi said that this work can dramatically reduce the hardware cost by replacing the neurons and synapses that were based on complex digital and analog circuits with a single transistor. "We have demonstrated that neurons and synapses can be implemented using a single transistor," said Joon-Kyu Han, the first author. "By co-integrating single transistor neurons and synapses on the same wafer using a standard CMOS process, the hardware cost of the neuromorphic hardware has been improved, which will accelerate the commercialization of neuromorphic hardware,” Han added.This research was supported by the National Research Foundation (NRF) and IC Design Education Center (IDEC).
-PublicationJoon-Kyu Han, Sung-Yool Choi, Yang-Kyu Choi, et al.“Cointegration of single-transistor neurons and synapses by nanoscale CMOS fabrication for highly scalable neuromorphic hardware,” Science Advances (DOI: 10.1126/sciadv.abg8836)
-ProfileProfessor Yang-Kyu ChoiNano-Oriented Bio-Electronics Labhttps://sites.google.com/view/nobelab/
School of Electrical EngineeringKAIST
Professor Sung-Yool ChoiMolecular and Nano Device Laboratoryhttps://www.mndl.kaist.ac.kr/
School of Electrical EngineeringKAIST
2021.08.05 View 7213
Brain-Inspired Highly Scalable Neuromorphic Hardware Presented
Neurons and synapses based on single transistor can dramatically reduce the hardware cost and accelerate the commercialization of neuromorphic hardware
KAIST researchers fabricated a brain-inspired highly scalable neuromorphic hardware by co-integrating single transistor neurons and synapses. Using standard silicon complementary metal-oxide-semiconductor (CMOS) technology, the neuromorphic hardware is expected to reduce chip cost and simplify fabrication procedures.
The research team led by Yang-Kyu Choi and Sung-Yool Choi produced a neurons and synapses based on single transistor for highly scalable neuromorphic hardware and showed the ability to recognize text and face images. This research was featured in Science Advances on August 4.
Neuromorphic hardware has attracted a great deal of attention because of its artificial intelligence functions, but consuming ultra-low power of less than 20 watts by mimicking the human brain. To make neuromorphic hardware work, a neuron that generates a spike when integrating a certain signal, and a synapse remembering the connection between two neurons are necessary, just like the biological brain. However, since neurons and synapses constructed on digital or analog circuits occupy a large space, there is a limit in terms of hardware efficiency and costs. Since the human brain consists of about 1011 neurons and 1014 synapses, it is necessary to improve the hardware cost in order to apply it to mobile and IoT devices.
To solve the problem, the research team mimicked the behavior of biological neurons and synapses with a single transistor, and co-integrated them onto an 8-inch wafer. The manufactured neuromorphic transistors have the same structure as the transistors for memory and logic that are currently mass-produced. In addition, the neuromorphic transistors proved for the first time that they can be implemented with a ‘Janus structure’ that functions as both neuron and synapse, just like coins have heads and tails.
Professor Yang-Kyu Choi said that this work can dramatically reduce the hardware cost by replacing the neurons and synapses that were based on complex digital and analog circuits with a single transistor. "We have demonstrated that neurons and synapses can be implemented using a single transistor," said Joon-Kyu Han, the first author. "By co-integrating single transistor neurons and synapses on the same wafer using a standard CMOS process, the hardware cost of the neuromorphic hardware has been improved, which will accelerate the commercialization of neuromorphic hardware,” Han added.This research was supported by the National Research Foundation (NRF) and IC Design Education Center (IDEC).
-PublicationJoon-Kyu Han, Sung-Yool Choi, Yang-Kyu Choi, et al.“Cointegration of single-transistor neurons and synapses by nanoscale CMOS fabrication for highly scalable neuromorphic hardware,” Science Advances (DOI: 10.1126/sciadv.abg8836)
-ProfileProfessor Yang-Kyu ChoiNano-Oriented Bio-Electronics Labhttps://sites.google.com/view/nobelab/
School of Electrical EngineeringKAIST
Professor Sung-Yool ChoiMolecular and Nano Device Laboratoryhttps://www.mndl.kaist.ac.kr/
School of Electrical EngineeringKAIST
2021.08.05 View 7213 -
 3D Visualization and Quantification of Bioplastic PHA in a Living Bacterial Cell
3D holographic microscopy leads to in-depth analysis of bacterial cells accumulating the bacterial bioplastic, polyhydroxyalkanoate (PHA)
A research team at KAIST has observed how bioplastic granule is being accumulated in living bacteria cells through 3D holographic microscopy. Their 3D imaging and quantitative analysis of the bioplastic ‘polyhydroxyalkanoate’ (PHA) via optical diffraction tomography provides insights into biosynthesizing sustainable substitutes for petroleum-based plastics.
The bio-degradable polyester polyhydroxyalkanoate (PHA) is being touted as an eco-friendly bioplastic to replace existing synthetic plastics. While carrying similar properties to general-purpose plastics such as polyethylene and polypropylene, PHA can be used in various industrial applications such as container packaging and disposable products.
PHA is synthesized by numerous bacteria as an energy and carbon storage material under unbalanced growth conditions in the presence of excess carbon sources. PHA exists in the form of insoluble granules in the cytoplasm. Previous studies on investigating in vivo PHA granules have been performed by using fluorescence microscopy, transmission electron microscopy (TEM), and electron cryotomography.
These techniques have generally relied on the statistical analysis of multiple 2D snapshots of fixed cells or the short-time monitoring of the cells. For the TEM analysis, cells need to be fixed and sectioned, and thus the investigation of living cells was not possible. Fluorescence-based techniques require fluorescence labeling or dye staining. Thus, indirect imaging with the use of reporter proteins cannot show the native state of PHAs or cells, and invasive exogenous dyes can affect the physiology and viability of the cells. Therefore, it was difficult to fully understand the formation of PHA granules in cells due to the technical limitations, and thus several mechanism models based on the observations have been only proposed.
The team of metabolic engineering researchers led by Distinguished Professor Sang Yup Lee and Physics Professor YongKeun Park, who established the startup Tomocube with his 3D holographic microscopy, reported the results of 3D quantitative label-free analysis of PHA granules in individual live bacterial cells by measuring the refractive index distributions using optical diffraction tomography. The formation and growth of PHA granules in the cells of Cupriavidus necator, the most-studied native PHA (specifically, poly(3-hydroxybutyrate), also known as PHB) producer, and recombinant Escherichia coli harboring C. necator PHB biosynthesis pathway were comparatively examined.
From the reconstructed 3D refractive index distribution of the cells, the team succeeded in the 3D visualization and quantitative analysis of cells and intracellular PHA granules at a single-cell level. In particular, the team newly presented the concept of “in vivo PHA granule density.” Through the statistical analysis of hundreds of single cells accumulating PHA granules, the distinctive differences of density and localization of PHA granules in the two micro-organisms were found. Furthermore, the team identified the key protein that plays a major role in making the difference that enabled the characteristics of PHA granules in the recombinant E. coli to become similar to those of C. necator.
The research team also presented 3D time-lapse movies showing the actual processes of PHA granule formation combined with cell growth and division. Movies showing the living cells synthesizing and accumulating PHA granules in their native state had never been reported before.
Professor Lee said, “This study provides insights into the morphological and physical characteristics of in vivo PHA as well as the unique mechanisms of PHA granule formation that undergo the phase transition from soluble monomers into the insoluble polymer, followed by granule formation. Through this study, a deeper understanding of PHA granule formation within the bacterial cells is now possible, which has great significance in that a convergence study of biology and physics was achieved. This study will help develop various bioplastics production processes in the future.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (Grants NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) and the Bio & Medical Technology Development Program (Grant No. 2021M3A9I4022740) from the Ministry of Science and ICT (MSIT) through the National Research Foundation (NRF) of Korea to S.Y.L. This work was also supported by the KAIST Cross-Generation Collaborative Laboratory project.
-PublicationSo Young Choi, Jeonghun Oh, JaeHwang Jung, YongKeun Park, and Sang Yup Lee. Three-dimensional label-free visualization and quantification of polyhydroxyalkanoates in individualbacterial cell in its native state. PNAS(https://doi.org./10.1073/pnas.2103956118)
-ProfileDistinguished Professor Sang Yup LeeMetabolic Engineering and Synthetic Biologyhttp://mbel.kaist.ac.kr/
Department of Chemical and Biomolecular Engineering KAIST
Endowed Chair Professor YongKeun ParkBiomedical Optics Laboratoryhttps://bmokaist.wordpress.com/
Department of PhysicsKAIST
2021.07.28 View 8229
3D Visualization and Quantification of Bioplastic PHA in a Living Bacterial Cell
3D holographic microscopy leads to in-depth analysis of bacterial cells accumulating the bacterial bioplastic, polyhydroxyalkanoate (PHA)
A research team at KAIST has observed how bioplastic granule is being accumulated in living bacteria cells through 3D holographic microscopy. Their 3D imaging and quantitative analysis of the bioplastic ‘polyhydroxyalkanoate’ (PHA) via optical diffraction tomography provides insights into biosynthesizing sustainable substitutes for petroleum-based plastics.
The bio-degradable polyester polyhydroxyalkanoate (PHA) is being touted as an eco-friendly bioplastic to replace existing synthetic plastics. While carrying similar properties to general-purpose plastics such as polyethylene and polypropylene, PHA can be used in various industrial applications such as container packaging and disposable products.
PHA is synthesized by numerous bacteria as an energy and carbon storage material under unbalanced growth conditions in the presence of excess carbon sources. PHA exists in the form of insoluble granules in the cytoplasm. Previous studies on investigating in vivo PHA granules have been performed by using fluorescence microscopy, transmission electron microscopy (TEM), and electron cryotomography.
These techniques have generally relied on the statistical analysis of multiple 2D snapshots of fixed cells or the short-time monitoring of the cells. For the TEM analysis, cells need to be fixed and sectioned, and thus the investigation of living cells was not possible. Fluorescence-based techniques require fluorescence labeling or dye staining. Thus, indirect imaging with the use of reporter proteins cannot show the native state of PHAs or cells, and invasive exogenous dyes can affect the physiology and viability of the cells. Therefore, it was difficult to fully understand the formation of PHA granules in cells due to the technical limitations, and thus several mechanism models based on the observations have been only proposed.
The team of metabolic engineering researchers led by Distinguished Professor Sang Yup Lee and Physics Professor YongKeun Park, who established the startup Tomocube with his 3D holographic microscopy, reported the results of 3D quantitative label-free analysis of PHA granules in individual live bacterial cells by measuring the refractive index distributions using optical diffraction tomography. The formation and growth of PHA granules in the cells of Cupriavidus necator, the most-studied native PHA (specifically, poly(3-hydroxybutyrate), also known as PHB) producer, and recombinant Escherichia coli harboring C. necator PHB biosynthesis pathway were comparatively examined.
From the reconstructed 3D refractive index distribution of the cells, the team succeeded in the 3D visualization and quantitative analysis of cells and intracellular PHA granules at a single-cell level. In particular, the team newly presented the concept of “in vivo PHA granule density.” Through the statistical analysis of hundreds of single cells accumulating PHA granules, the distinctive differences of density and localization of PHA granules in the two micro-organisms were found. Furthermore, the team identified the key protein that plays a major role in making the difference that enabled the characteristics of PHA granules in the recombinant E. coli to become similar to those of C. necator.
The research team also presented 3D time-lapse movies showing the actual processes of PHA granule formation combined with cell growth and division. Movies showing the living cells synthesizing and accumulating PHA granules in their native state had never been reported before.
Professor Lee said, “This study provides insights into the morphological and physical characteristics of in vivo PHA as well as the unique mechanisms of PHA granule formation that undergo the phase transition from soluble monomers into the insoluble polymer, followed by granule formation. Through this study, a deeper understanding of PHA granule formation within the bacterial cells is now possible, which has great significance in that a convergence study of biology and physics was achieved. This study will help develop various bioplastics production processes in the future.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (Grants NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) and the Bio & Medical Technology Development Program (Grant No. 2021M3A9I4022740) from the Ministry of Science and ICT (MSIT) through the National Research Foundation (NRF) of Korea to S.Y.L. This work was also supported by the KAIST Cross-Generation Collaborative Laboratory project.
-PublicationSo Young Choi, Jeonghun Oh, JaeHwang Jung, YongKeun Park, and Sang Yup Lee. Three-dimensional label-free visualization and quantification of polyhydroxyalkanoates in individualbacterial cell in its native state. PNAS(https://doi.org./10.1073/pnas.2103956118)
-ProfileDistinguished Professor Sang Yup LeeMetabolic Engineering and Synthetic Biologyhttp://mbel.kaist.ac.kr/
Department of Chemical and Biomolecular Engineering KAIST
Endowed Chair Professor YongKeun ParkBiomedical Optics Laboratoryhttps://bmokaist.wordpress.com/
Department of PhysicsKAIST
2021.07.28 View 8229 -
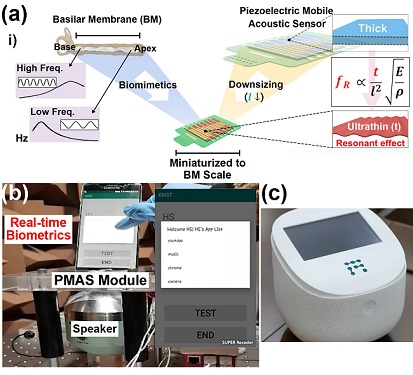 Biomimetic Resonant Acoustic Sensor Detecting Far-Distant Voices Accurately to Hit the Market
A KAIST research team led by Professor Keon Jae Lee from the Department of Materials Science and Engineering has developed a bioinspired flexible piezoelectric acoustic sensor with multi-resonant ultrathin piezoelectric membrane mimicking the basilar membrane of the human cochlea. The flexible acoustic sensor has been miniaturized for embedding into smartphones and the first commercial prototype is ready for accurate and far-distant voice detection.
In 2018, Professor Lee presented the first concept of a flexible piezoelectric acoustic sensor, inspired by the fact that humans can accurately detect far-distant voices using a multi-resonant trapezoidal membrane with 20,000 hair cells. However, previous acoustic sensors could not be integrated into commercial products like smartphones and AI speakers due to their large device size.
In this work, the research team fabricated a mobile-sized acoustic sensor by adopting ultrathin piezoelectric membranes with high sensitivity. Simulation studies proved that the ultrathin polymer underneath inorganic piezoelectric thin film can broaden the resonant bandwidth to cover the entire voice frequency range using seven channels. Based on this theory, the research team successfully demonstrated the miniaturized acoustic sensor mounted in commercial smartphones and AI speakers for machine learning-based biometric authentication and voice processing. (Please refer to the explanatory movie KAIST Flexible Piezoelectric Mobile Acoustic Sensor).
The resonant mobile acoustic sensor has superior sensitivity and multi-channel signals compared to conventional condenser microphones with a single channel, and it has shown highly accurate and far-distant speaker identification with a small amount of voice training data. The error rate of speaker identification was significantly reduced by 56% (with 150 training datasets) and 75% (with 2,800 training datasets) compared to that of a MEMS condenser device.
Professor Lee said, “Recently, Google has been targeting the ‘Wolverine Project’ on far-distant voice separation from multi-users for next-generation AI user interfaces. I expect that our multi-channel resonant acoustic sensor with abundant voice information is the best fit for this application. Currently, the mass production process is on the verge of completion, so we hope that this will be used in our daily lives very soon.”
Professor Lee also established a startup company called Fronics Inc., located both in Korea and U.S. (branch office) to commercialize this flexible acoustic sensor and is seeking collaborations with global AI companies.
These research results entitled “Biomimetic and Flexible Piezoelectric Mobile Acoustic Sensors with Multi-Resonant Ultrathin Structures for Machine Learning Biometrics” were published in Science Advances in 2021 (7, eabe5683).
-Publication
“Biomimetic and flexible piezoelectric mobile acoustic sensors with multiresonant ultrathin structures for machine learning biometrics,” Science Advances (DOI: 10.1126/sciadv.abe5683)
-Profile
Professor Keon Jae Lee
Department of Materials Science and Engineering
Flexible and Nanobio Device Lab
http://fand.kaist.ac.kr/
KAIST
2021.06.14 View 6934
Biomimetic Resonant Acoustic Sensor Detecting Far-Distant Voices Accurately to Hit the Market
A KAIST research team led by Professor Keon Jae Lee from the Department of Materials Science and Engineering has developed a bioinspired flexible piezoelectric acoustic sensor with multi-resonant ultrathin piezoelectric membrane mimicking the basilar membrane of the human cochlea. The flexible acoustic sensor has been miniaturized for embedding into smartphones and the first commercial prototype is ready for accurate and far-distant voice detection.
In 2018, Professor Lee presented the first concept of a flexible piezoelectric acoustic sensor, inspired by the fact that humans can accurately detect far-distant voices using a multi-resonant trapezoidal membrane with 20,000 hair cells. However, previous acoustic sensors could not be integrated into commercial products like smartphones and AI speakers due to their large device size.
In this work, the research team fabricated a mobile-sized acoustic sensor by adopting ultrathin piezoelectric membranes with high sensitivity. Simulation studies proved that the ultrathin polymer underneath inorganic piezoelectric thin film can broaden the resonant bandwidth to cover the entire voice frequency range using seven channels. Based on this theory, the research team successfully demonstrated the miniaturized acoustic sensor mounted in commercial smartphones and AI speakers for machine learning-based biometric authentication and voice processing. (Please refer to the explanatory movie KAIST Flexible Piezoelectric Mobile Acoustic Sensor).
The resonant mobile acoustic sensor has superior sensitivity and multi-channel signals compared to conventional condenser microphones with a single channel, and it has shown highly accurate and far-distant speaker identification with a small amount of voice training data. The error rate of speaker identification was significantly reduced by 56% (with 150 training datasets) and 75% (with 2,800 training datasets) compared to that of a MEMS condenser device.
Professor Lee said, “Recently, Google has been targeting the ‘Wolverine Project’ on far-distant voice separation from multi-users for next-generation AI user interfaces. I expect that our multi-channel resonant acoustic sensor with abundant voice information is the best fit for this application. Currently, the mass production process is on the verge of completion, so we hope that this will be used in our daily lives very soon.”
Professor Lee also established a startup company called Fronics Inc., located both in Korea and U.S. (branch office) to commercialize this flexible acoustic sensor and is seeking collaborations with global AI companies.
These research results entitled “Biomimetic and Flexible Piezoelectric Mobile Acoustic Sensors with Multi-Resonant Ultrathin Structures for Machine Learning Biometrics” were published in Science Advances in 2021 (7, eabe5683).
-Publication
“Biomimetic and flexible piezoelectric mobile acoustic sensors with multiresonant ultrathin structures for machine learning biometrics,” Science Advances (DOI: 10.1126/sciadv.abe5683)
-Profile
Professor Keon Jae Lee
Department of Materials Science and Engineering
Flexible and Nanobio Device Lab
http://fand.kaist.ac.kr/
KAIST
2021.06.14 View 6934 -
 Microbial Production of a Natural Red Colorant Carminic Acid
Metabolic engineering and computer-simulated enzyme engineering led to the production of carminic acid, a natural red colorant, from bacteria for the first time
A research group at KAIST has engineered a bacterium capable of producing a natural red colorant, carminic acid, which is widely used for food and cosmetics. The research team reported the complete biosynthesis of carminic acid from glucose in engineered Escherichia coli. The strategies will be useful for the design and construction of biosynthetic pathways involving unknown enzymes and consequently the production of diverse industrially important natural products for the food, pharmaceutical, and cosmetic industries.
Carminic acid is a natural red colorant widely being used for products such as strawberry milk and lipstick. However, carminic acid has been produced by farming cochineals, a scale insect which only grows in the region around Peru and Canary Islands, followed by complicated multi-step purification processes. Moreover, carminic acid often contains protein contaminants that cause allergies so many people are unwilling to consume products made of insect-driven colorants. On that account, manufacturers around the world are using alternative red colorants despite the fact that carminic acid is one of the most stable natural red colorants.
These challenges inspired the metabolic engineering research group at KAIST to address this issue. Its members include postdoctoral researchers Dongsoo Yang and Woo Dae Jang, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering. This study entitled “Production of carminic acid by metabolically engineered Escherichia coli” was published online in the Journal of the American Chemical Society (JACS) on April 2.
This research reports for the first time the development of a bacterial strain capable of producing carminic acid from glucose via metabolic engineering and computer simulation-assisted enzyme engineering. The research group optimized the type II polyketide synthase machinery to efficiently produce the precursor of carminic acid, flavokermesic acid.
Since the enzymes responsible for the remaining two reactions were neither discovered nor functional, biochemical reaction analysis was performed to identify enzymes that can convert flavokermesic acid into carminic acid. Then, homology modeling and docking simulations were performed to enhance the activities of the two identified enzymes. The team could confirm that the final engineered strain could produce carminic acid directly from glucose. The C-glucosyltransferase developed in this study was found to be generally applicable for other natural products as showcased by the successful production of an additional product, aloesin, which is found in aloe leaves.
“The most important part of this research is that unknown enzymes for the production of target natural products were identified and improved by biochemical reaction analyses and computer simulation-assisted enzyme engineering,” says Dr. Dongsoo Yang. He explained the development of a generally applicable C-glucosyltransferase is also useful since C-glucosylation is a relatively unexplored reaction in bacteria including Escherichia coli. Using the C-glucosyltransferase developed in this study, both carminic acid and aloesin were successfully produced from glucose.
“A sustainable and insect-free method of producing carminic acid was achieved for the first time in this study. Unknown or inefficient enzymes have always been a major problem in natural product biosynthesis, and here we suggest one effective solution for solving this problem. As maintaining good health in the aging society is becoming increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other valuable natural products of medical or nutritional importance,” said Distinguished Professor Sang Yup Lee.
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries of the Ministry of Science and ICT (MSIT) through the National Research Foundation (NRF) of Korea and the KAIST Cross-Generation Collaborative Lab project; Sang Yup Lee and Dongsoo Yang were also supported by Novo Nordisk Foundation in Denmark.
Publication:
Dongsoo Yang, Woo Dae Jang, and Sang Yup Lee. Production of carminic acid by metabolically engineered Escherichia coli. at the Journal of the American Chemical Society. https://doi.org.10.1021/jacs.0c12406
Profile:
Sang Yup Lee, PhD
Distinguished Professor
leesy@kaist.ac.kr
http://mbel.kaist.ac.kr
Metabolic &Biomolecular Engineering National Research Laboratory
Department of Chemical and Biomolecular Engineering
KAIST
2021.04.06 View 8420
Microbial Production of a Natural Red Colorant Carminic Acid
Metabolic engineering and computer-simulated enzyme engineering led to the production of carminic acid, a natural red colorant, from bacteria for the first time
A research group at KAIST has engineered a bacterium capable of producing a natural red colorant, carminic acid, which is widely used for food and cosmetics. The research team reported the complete biosynthesis of carminic acid from glucose in engineered Escherichia coli. The strategies will be useful for the design and construction of biosynthetic pathways involving unknown enzymes and consequently the production of diverse industrially important natural products for the food, pharmaceutical, and cosmetic industries.
Carminic acid is a natural red colorant widely being used for products such as strawberry milk and lipstick. However, carminic acid has been produced by farming cochineals, a scale insect which only grows in the region around Peru and Canary Islands, followed by complicated multi-step purification processes. Moreover, carminic acid often contains protein contaminants that cause allergies so many people are unwilling to consume products made of insect-driven colorants. On that account, manufacturers around the world are using alternative red colorants despite the fact that carminic acid is one of the most stable natural red colorants.
These challenges inspired the metabolic engineering research group at KAIST to address this issue. Its members include postdoctoral researchers Dongsoo Yang and Woo Dae Jang, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering. This study entitled “Production of carminic acid by metabolically engineered Escherichia coli” was published online in the Journal of the American Chemical Society (JACS) on April 2.
This research reports for the first time the development of a bacterial strain capable of producing carminic acid from glucose via metabolic engineering and computer simulation-assisted enzyme engineering. The research group optimized the type II polyketide synthase machinery to efficiently produce the precursor of carminic acid, flavokermesic acid.
Since the enzymes responsible for the remaining two reactions were neither discovered nor functional, biochemical reaction analysis was performed to identify enzymes that can convert flavokermesic acid into carminic acid. Then, homology modeling and docking simulations were performed to enhance the activities of the two identified enzymes. The team could confirm that the final engineered strain could produce carminic acid directly from glucose. The C-glucosyltransferase developed in this study was found to be generally applicable for other natural products as showcased by the successful production of an additional product, aloesin, which is found in aloe leaves.
“The most important part of this research is that unknown enzymes for the production of target natural products were identified and improved by biochemical reaction analyses and computer simulation-assisted enzyme engineering,” says Dr. Dongsoo Yang. He explained the development of a generally applicable C-glucosyltransferase is also useful since C-glucosylation is a relatively unexplored reaction in bacteria including Escherichia coli. Using the C-glucosyltransferase developed in this study, both carminic acid and aloesin were successfully produced from glucose.
“A sustainable and insect-free method of producing carminic acid was achieved for the first time in this study. Unknown or inefficient enzymes have always been a major problem in natural product biosynthesis, and here we suggest one effective solution for solving this problem. As maintaining good health in the aging society is becoming increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other valuable natural products of medical or nutritional importance,” said Distinguished Professor Sang Yup Lee.
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries of the Ministry of Science and ICT (MSIT) through the National Research Foundation (NRF) of Korea and the KAIST Cross-Generation Collaborative Lab project; Sang Yup Lee and Dongsoo Yang were also supported by Novo Nordisk Foundation in Denmark.
Publication:
Dongsoo Yang, Woo Dae Jang, and Sang Yup Lee. Production of carminic acid by metabolically engineered Escherichia coli. at the Journal of the American Chemical Society. https://doi.org.10.1021/jacs.0c12406
Profile:
Sang Yup Lee, PhD
Distinguished Professor
leesy@kaist.ac.kr
http://mbel.kaist.ac.kr
Metabolic &Biomolecular Engineering National Research Laboratory
Department of Chemical and Biomolecular Engineering
KAIST
2021.04.06 View 8420 -
 Professor Jae Kyoung Kim to Lead a New Mathematical Biology Research Group at IBS
Professor Jae Kyoung Kim from the KAIST Department of Mathematical Sciences was appointed as the third Chief Investigator (CI) of the Pioneer Research Center (PRC) for Mathematical and Computational Sciences at the Institute for Basic Science (IBS). Professor Kim will launch and lead a new research group that will be devoted to resolving various biological conundrums from a mathematical perspective. His appointment began on March 1, 2021.
Professor Kim, a rising researcher in the field of mathematical biology, has received attention from both the mathematical and biological communities at the international level. Professor Kim puts novel and unremitting efforts into understanding biological systems such as cell-to-cell interactions mathematically and designing mathematical models for identifying causes of diseases and developing therapeutic medicines.
Through active joint research with biologists, mathematician Kim has addressed many challenges that have remained unsolved in biology and published papers in a number of leading international journals in related fields. His notable works based on mathematical modelling include having designed a biological circuit that can maintain a stable circadian rhythm (Science, 2015) and unveiling the principles of how the biological clock in the body maintains a steady speed for the first time in over 60 years (Molecular Cell, 2015). Recently, through a joint research project with Pfizer, Professor Kim identified what causes the differences between animal and clinical test results during drug development explaining why drugs have different efficacies in different people (Molecular Systems Biology, 2019).
The new IBS biomedical mathematics research group led by Professor Kim will further investigate the causes of unstable circadian rhythms and sleeping patterns. The team will aim to present a new paradigm in treatments for sleep disorders.
Professor Kim said, “We are all so familiar with sleep behaviors, but the exact mechanisms behind how such behaviors occur are still unknown. Through cooperation with biomedical scientists, our group will do its best to discover the complicated, fundamental mechanisms of sleep, and investigate the causes and cures of sleep disorders.”
Every year, the IBS selects young and promising researchers and appoints them as CIs. A maximum of five selected CIs can form each independent research group within the IBS PRC, and receive research funds of 1 billion to 1.5 billion KRW over five years.
(END)
2021.03.18 View 6611
Professor Jae Kyoung Kim to Lead a New Mathematical Biology Research Group at IBS
Professor Jae Kyoung Kim from the KAIST Department of Mathematical Sciences was appointed as the third Chief Investigator (CI) of the Pioneer Research Center (PRC) for Mathematical and Computational Sciences at the Institute for Basic Science (IBS). Professor Kim will launch and lead a new research group that will be devoted to resolving various biological conundrums from a mathematical perspective. His appointment began on March 1, 2021.
Professor Kim, a rising researcher in the field of mathematical biology, has received attention from both the mathematical and biological communities at the international level. Professor Kim puts novel and unremitting efforts into understanding biological systems such as cell-to-cell interactions mathematically and designing mathematical models for identifying causes of diseases and developing therapeutic medicines.
Through active joint research with biologists, mathematician Kim has addressed many challenges that have remained unsolved in biology and published papers in a number of leading international journals in related fields. His notable works based on mathematical modelling include having designed a biological circuit that can maintain a stable circadian rhythm (Science, 2015) and unveiling the principles of how the biological clock in the body maintains a steady speed for the first time in over 60 years (Molecular Cell, 2015). Recently, through a joint research project with Pfizer, Professor Kim identified what causes the differences between animal and clinical test results during drug development explaining why drugs have different efficacies in different people (Molecular Systems Biology, 2019).
The new IBS biomedical mathematics research group led by Professor Kim will further investigate the causes of unstable circadian rhythms and sleeping patterns. The team will aim to present a new paradigm in treatments for sleep disorders.
Professor Kim said, “We are all so familiar with sleep behaviors, but the exact mechanisms behind how such behaviors occur are still unknown. Through cooperation with biomedical scientists, our group will do its best to discover the complicated, fundamental mechanisms of sleep, and investigate the causes and cures of sleep disorders.”
Every year, the IBS selects young and promising researchers and appoints them as CIs. A maximum of five selected CIs can form each independent research group within the IBS PRC, and receive research funds of 1 billion to 1.5 billion KRW over five years.
(END)
2021.03.18 View 6611