Sang+Yup+Lee
-
 Distinguished Professor Sang Yup Lee Attends World Economic Forum's Workshop
Sang Yup Lee, Distinguished Professor of Chemical and Biomolecular Engineering at KAIST was invited to attend the Technology Pioneer and Global Growth Company CEO Workshop hosted by the World Economic Forum (WEF) on June 19-20, 2014 in San Francisco. During the workshop, Professor Lee joined a discussion on “disruptive technologies” as a panelist.
Currently serving for the evaluation committee that selects technology pioneers for the workshop, during the discussion, Professor Lee identified important issues facing humanity, analyzed the issues through forecasting, and presented converging disruptive technologies that provide solutions to the problems. He also shared the “ten emerging technologies” announced by the Global Agenda Council on Emerging Technologies, WEF and the Korean government’s technology innovation strategies adopted to achieve its economic development policy called creative economy.
2014.06.22 View 6796
Distinguished Professor Sang Yup Lee Attends World Economic Forum's Workshop
Sang Yup Lee, Distinguished Professor of Chemical and Biomolecular Engineering at KAIST was invited to attend the Technology Pioneer and Global Growth Company CEO Workshop hosted by the World Economic Forum (WEF) on June 19-20, 2014 in San Francisco. During the workshop, Professor Lee joined a discussion on “disruptive technologies” as a panelist.
Currently serving for the evaluation committee that selects technology pioneers for the workshop, during the discussion, Professor Lee identified important issues facing humanity, analyzed the issues through forecasting, and presented converging disruptive technologies that provide solutions to the problems. He also shared the “ten emerging technologies” announced by the Global Agenda Council on Emerging Technologies, WEF and the Korean government’s technology innovation strategies adopted to achieve its economic development policy called creative economy.
2014.06.22 View 6796 -
 KAIST Participates in the 2014 Davos Forum on January 22-25 in Switzerland
Through the sessions of the Global University Leaders Forum, IdeasLab, and Global Agenda Councils on Biotechnology, KAIST participants will actively engage with global leaders in the discussion of issues on education innovation and technological breakthroughs.
The 2014 Annual Meeting of the World Economic Forum (WEF), known as the Davos Forum, will kick off on January 22-25 in Davos-Klosters, Switzerland, under the theme of "The Reshaping of the World: Consequences for Society, Politics, and Business." Each year, the Forum attracts about 2,500 distinguished leaders from all around the world and provides an open platform to identify the current and emerging challenges facing the global community and to develop ideas and actions necessary to respond to such challenges.
President Sung-Mo Steve Kang and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering, KAIST, will attend the Forum and engage in a series of dialogues on such issues as Massive Open Online Courses, new paradigms for universities and researchers, the transformation of higher education, the role and value of scientific discoveries, and the impact of biotechnology on the future of society and business.
At the session entitled "New Paradigms for Universities of the Future" hosted by the Global University Leaders Forum (GULF), President Kang will introduce KAIST"s ongoing online education program, Education 3.0. GULF was created in 2006 by WEF, which is a small community of the presidents and senior representatives of the top universities in the world.
Implemented in 2012, Education 3.0 incorporates advanced information and communications technology (ICT) to offer students and teachers a learner-based, team-oriented learning and teaching environment. Under Education 3.0, students study online and meet in groups with a professor for in-depth discussions, collaboration, and problem-solving. KAIST plans to expand the program to embrace the global community in earnest by establishing Education 3.0 Global in order to have interactive real-time classes for students and researchers across regions and cultures.
President Kang will also present a paper entitled "Toward Socially Responsible Technology: KAIST"s Approach to Integrating Social and Behavioral Perspectives into Technology Development" at another session of GULF called "Seeking New Approaches to Critical Global Challenges." In the paper, President Kang points out that notwithstanding the many benefits we enjoy from the increasingly interconnected world, digital media may pose a threat to become a new outlet for social problems, for example, Internet or digital addiction.
Experts say that early exposure to digital devices harms the healthy development of cognitive functions, emotions, and social behavior. President Kang will introduce KAIST"s recent endeavor to develop a non-intrusive technology to help prevent digital addiction, which will ultimately be embedded in the form of a virtual coach or mentor that helps and guides people under risk to make constructive use of digital devices. President Kang stresses the fundamental shift in the science and technology development paradigm from research and development (R&D) to a research and solution development (R&SD), taking serious consideration of societal needs, quality of life, and social impacts when conducting research.
Professor Sang Yup Lee will moderate the IdeasLab session at the Davos Forum entitled "From Lab to Life with the California Institute of Technology (Caltech)." Together with scientists from Caltech, he will discuss scientific breakthroughs that transform institutions, industries, and individuals in the near future, such as the development of damage-tolerant lightweight materials with nanotechnology, the ability to read and write genomes, and wireless lab-in-the-body monitors. In addition, he will meet global business leaders at the session of "Sustainability, Innovation, and Growth" and speak about how emerging technologies, biotechnology in particular, will transform future societies, business, and industries.
As a current special adviser of the World Economic Forum"s (WEF) Chemicals Industry Community, Professor Lee will meet global chairs and chief executive officers of chemical companies and discuss ways to advance the industry to become more bio-based and environmentally friendly. He served as a founding chairman of WEF"s Global Agenda Councils on Biotechnology in 2013.
President Sung-Mo Steve Kang Distinguished Professor Sang Yup Lee
2014.01.17 View 8472
KAIST Participates in the 2014 Davos Forum on January 22-25 in Switzerland
Through the sessions of the Global University Leaders Forum, IdeasLab, and Global Agenda Councils on Biotechnology, KAIST participants will actively engage with global leaders in the discussion of issues on education innovation and technological breakthroughs.
The 2014 Annual Meeting of the World Economic Forum (WEF), known as the Davos Forum, will kick off on January 22-25 in Davos-Klosters, Switzerland, under the theme of "The Reshaping of the World: Consequences for Society, Politics, and Business." Each year, the Forum attracts about 2,500 distinguished leaders from all around the world and provides an open platform to identify the current and emerging challenges facing the global community and to develop ideas and actions necessary to respond to such challenges.
President Sung-Mo Steve Kang and Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering, KAIST, will attend the Forum and engage in a series of dialogues on such issues as Massive Open Online Courses, new paradigms for universities and researchers, the transformation of higher education, the role and value of scientific discoveries, and the impact of biotechnology on the future of society and business.
At the session entitled "New Paradigms for Universities of the Future" hosted by the Global University Leaders Forum (GULF), President Kang will introduce KAIST"s ongoing online education program, Education 3.0. GULF was created in 2006 by WEF, which is a small community of the presidents and senior representatives of the top universities in the world.
Implemented in 2012, Education 3.0 incorporates advanced information and communications technology (ICT) to offer students and teachers a learner-based, team-oriented learning and teaching environment. Under Education 3.0, students study online and meet in groups with a professor for in-depth discussions, collaboration, and problem-solving. KAIST plans to expand the program to embrace the global community in earnest by establishing Education 3.0 Global in order to have interactive real-time classes for students and researchers across regions and cultures.
President Kang will also present a paper entitled "Toward Socially Responsible Technology: KAIST"s Approach to Integrating Social and Behavioral Perspectives into Technology Development" at another session of GULF called "Seeking New Approaches to Critical Global Challenges." In the paper, President Kang points out that notwithstanding the many benefits we enjoy from the increasingly interconnected world, digital media may pose a threat to become a new outlet for social problems, for example, Internet or digital addiction.
Experts say that early exposure to digital devices harms the healthy development of cognitive functions, emotions, and social behavior. President Kang will introduce KAIST"s recent endeavor to develop a non-intrusive technology to help prevent digital addiction, which will ultimately be embedded in the form of a virtual coach or mentor that helps and guides people under risk to make constructive use of digital devices. President Kang stresses the fundamental shift in the science and technology development paradigm from research and development (R&D) to a research and solution development (R&SD), taking serious consideration of societal needs, quality of life, and social impacts when conducting research.
Professor Sang Yup Lee will moderate the IdeasLab session at the Davos Forum entitled "From Lab to Life with the California Institute of Technology (Caltech)." Together with scientists from Caltech, he will discuss scientific breakthroughs that transform institutions, industries, and individuals in the near future, such as the development of damage-tolerant lightweight materials with nanotechnology, the ability to read and write genomes, and wireless lab-in-the-body monitors. In addition, he will meet global business leaders at the session of "Sustainability, Innovation, and Growth" and speak about how emerging technologies, biotechnology in particular, will transform future societies, business, and industries.
As a current special adviser of the World Economic Forum"s (WEF) Chemicals Industry Community, Professor Lee will meet global chairs and chief executive officers of chemical companies and discuss ways to advance the industry to become more bio-based and environmentally friendly. He served as a founding chairman of WEF"s Global Agenda Councils on Biotechnology in 2013.
President Sung-Mo Steve Kang Distinguished Professor Sang Yup Lee
2014.01.17 View 8472 -
 The World Economic Forum Invites KAIST to 2014 Davos Forum
President Steve Kang and Distinguished Professor Sang Yup Lee have been invited by the World Economic Forum (WEF) to attend its annual meeting slated for January 22-25, 2014 in Davos-Klosters, Switzerland.
The president will also join the Global University Leaders Forum (GULF) to be held during the annual meeting. The GULF consists of leading research universities throughout the world, at which President Kang will address agenda related to higher education and research.
From September 11th to 13th, KAIST was invited to the WEF’s 2013 Summer Davos Forum held in Dalian, China. The Summer Davos Forum is recognized as a barometer of the world economy, and KAIST hosted three sessions there.
In a session titled “Smart Regulations,” Professor Sang Yup Lee hosted presentations and discussions under the topic of “How regulation models can strengthen technical innovation and expansion.” President Steve Kang, Peter Sands, CEO of Standard Chartered Bank Group, Mark Weinberger, CEO of Ernest & Young, and Peter Terium, CEO of RWE, participated in the discussions.
The KAIST delegates also presented and participated in a session titled “From Trade Center to Innovative Hub” to discuss how to lead innovations in Asia, as well as “Marine Resources: Finding New Frontier” to address issues of how to develop and manage oceanic resources for potential growth.
President Kang said, “The World Economic Forum allows us to introduce the results of our innovative and creative research to global leaders and to demonstrate that our global position continues to grow.”
The WEF has been hosting Summer Davos Forum in China since 2007. About 1,500 participants from over 90 countries joined in this year’s summer forum under the theme of “Innovation: Inevitable Mainstream.” New strategies for innovations and solutions for global threats were suggested through presentations and discussions in 125 sessions.
The World Economic Forum (WEF) is an independent, international, and non-profit organization based in Geneva, Switzerland. It is committed to improving the state of the world by engaging business, political, academic, and government leaders to shape global, regional and industry agenda.
Among the meetings and forums organized by the WEF, its annual meeting held each January in Davos, a.k.a. the Davos Forum, has been the best known gathering. The Davos Forum brings together some 2,500 top business leaders, international political leaders, selected intellectuals and journalists to discuss the most pressing issues facing the world including health and environment.
2013.11.07 View 8786
The World Economic Forum Invites KAIST to 2014 Davos Forum
President Steve Kang and Distinguished Professor Sang Yup Lee have been invited by the World Economic Forum (WEF) to attend its annual meeting slated for January 22-25, 2014 in Davos-Klosters, Switzerland.
The president will also join the Global University Leaders Forum (GULF) to be held during the annual meeting. The GULF consists of leading research universities throughout the world, at which President Kang will address agenda related to higher education and research.
From September 11th to 13th, KAIST was invited to the WEF’s 2013 Summer Davos Forum held in Dalian, China. The Summer Davos Forum is recognized as a barometer of the world economy, and KAIST hosted three sessions there.
In a session titled “Smart Regulations,” Professor Sang Yup Lee hosted presentations and discussions under the topic of “How regulation models can strengthen technical innovation and expansion.” President Steve Kang, Peter Sands, CEO of Standard Chartered Bank Group, Mark Weinberger, CEO of Ernest & Young, and Peter Terium, CEO of RWE, participated in the discussions.
The KAIST delegates also presented and participated in a session titled “From Trade Center to Innovative Hub” to discuss how to lead innovations in Asia, as well as “Marine Resources: Finding New Frontier” to address issues of how to develop and manage oceanic resources for potential growth.
President Kang said, “The World Economic Forum allows us to introduce the results of our innovative and creative research to global leaders and to demonstrate that our global position continues to grow.”
The WEF has been hosting Summer Davos Forum in China since 2007. About 1,500 participants from over 90 countries joined in this year’s summer forum under the theme of “Innovation: Inevitable Mainstream.” New strategies for innovations and solutions for global threats were suggested through presentations and discussions in 125 sessions.
The World Economic Forum (WEF) is an independent, international, and non-profit organization based in Geneva, Switzerland. It is committed to improving the state of the world by engaging business, political, academic, and government leaders to shape global, regional and industry agenda.
Among the meetings and forums organized by the WEF, its annual meeting held each January in Davos, a.k.a. the Davos Forum, has been the best known gathering. The Davos Forum brings together some 2,500 top business leaders, international political leaders, selected intellectuals and journalists to discuss the most pressing issues facing the world including health and environment.
2013.11.07 View 8786 -
 Professor Jae-Hyung Lee appointed as AIChE fellow
Professor Jae-Hyung Lee from the Department of Chemical and Bimolecular Engineering at KAIST was appointed as a fellow in the American Institute of Chemical Engineers (AIChE).
Established in 1908, AIChE is the largest association of chemical engineers worldwide, which now boasts more than 40,000 members from 90 countries.
Following Distinguished Professor Sang Yup Lee from the same department at KAIST, Professor Jae-Hyung Lee is the second Korean appointed as a fellow by the organization.
He has been acknowledged for his innovative research on the improvement of model predictive control of industrial processes.
Professor Lee is the director of the Saudi Armaco-KAIST CO2 Management Center at KAIST, a fellow of the Institute of Electrical and Electronics Engineers (IEEE) and the International Federation of Automatic Control (IFAC), and a member of the Korean Academy of Science and Technology.
He received the Young Investigator Award from the National Science Foundation (NSF) in 1994 and the Computing in Chemical Engineering Award from AIChE in 2013.
2013.11.05 View 7528
Professor Jae-Hyung Lee appointed as AIChE fellow
Professor Jae-Hyung Lee from the Department of Chemical and Bimolecular Engineering at KAIST was appointed as a fellow in the American Institute of Chemical Engineers (AIChE).
Established in 1908, AIChE is the largest association of chemical engineers worldwide, which now boasts more than 40,000 members from 90 countries.
Following Distinguished Professor Sang Yup Lee from the same department at KAIST, Professor Jae-Hyung Lee is the second Korean appointed as a fellow by the organization.
He has been acknowledged for his innovative research on the improvement of model predictive control of industrial processes.
Professor Lee is the director of the Saudi Armaco-KAIST CO2 Management Center at KAIST, a fellow of the Institute of Electrical and Electronics Engineers (IEEE) and the International Federation of Automatic Control (IFAC), and a member of the Korean Academy of Science and Technology.
He received the Young Investigator Award from the National Science Foundation (NSF) in 1994 and the Computing in Chemical Engineering Award from AIChE in 2013.
2013.11.05 View 7528 -
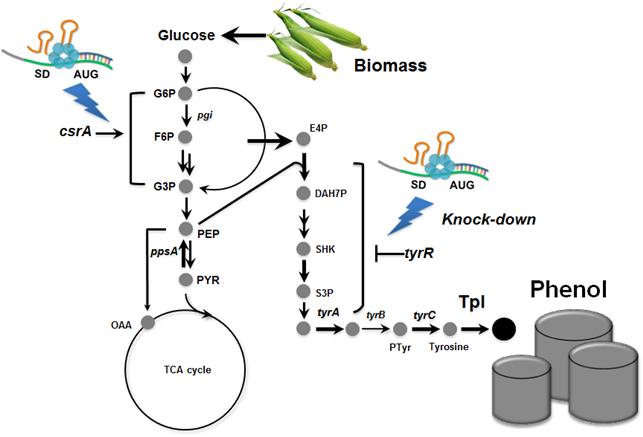 Metabolically engineered E. coli producing phenol
Many chemicals we use in everyday life are derived from fossil resources. Due to the increasing concerns on the use of fossil resources, there has been much interest in producing chemicals from renewable resources through biotechnology.
Phenol is an important commodity chemical, and is a starting material for the production of numerous industrial chemicals and polymers, including bisphenol A and phenolic resins, and others. At present, the production of phenol entirely depends on the chemical synthesis from benzene, and its annual production exceeds 8 million tons worldwide. Microbial production of phenol seems to be a non-viable process considering the high toxicity of phenol to the cell.
In the paper published online in Biotechnology Journal, a Korean research team led by Distinguished Professor Sang Yup Lee at the Department of Chemical and Biomolecular Engineering from the Korea Advanced Institute of Science and Technology (KAIST) reported the successful development of an engineered Escherichia coli (E. coli) strain which can produce phenol from glucose. E. coli has been a workhorse for biological production of various value-added compounds such as succinic acid and 1,4-butanediol in industrial scale. However, due to its low tolerance to phenol, E. coli was not considered a viable host strain for the biological production of phenol.
Professor Lee"s team, a leading research group in metabolic engineering, noted the genetic and physiological differences of various E. coli strains and investigated 18 different E. coli strains with respect to phenol tolerance and engineered all of the 18 strains simultaneously. If the traditional genetic engineering methods were used, this work would have taken years to do. To overcome this challenge, the research team used synthetic small RNA (sRNA) technology they recently developed (Nature Biotechnology, vol 31, pp 170-174, 2013). The sRNA technology allowed the team to screen 18 E. coli strains with respect to the phenol tolerance, and the activities of the metabolic pathway and enzyme involved in the production of phenol. The research team also metabolically engineered the E. coli strains to increase carbon flux toward phenol and finally generated an engineered E. coli strain which can produce phenol from glucose.
Furthermore, the team developed a biphasic extractive fermentation process to minimize the toxicity of phenol to E. coli cells. Glycerol tributyrate was found to have low toxicity to E. coli and allowed efficient extraction of phenol from the culture broth. Through the biphasic fed-batch fermentation using glycerol tributyrate as an in situ extractant, the final engineered E. coli strain produced phenol to the highest titer and productivity reported (3.8 g/L and 0.18 g/L/h, respectively). The strategy used for the strain development and the fermentation process will serve as a framework for metabolic engineering of microorganisms for the production of toxic chemicals from renewable resources.
This work was supported by the Intelligent Synthetic Biology Center through the Global Frontier Project (2011-0031963) of the Ministry of Science, ICT & Future Planning through the National Research Foundation of Korea.
Process of Phenol Production
2013.11.05 View 8074
Metabolically engineered E. coli producing phenol
Many chemicals we use in everyday life are derived from fossil resources. Due to the increasing concerns on the use of fossil resources, there has been much interest in producing chemicals from renewable resources through biotechnology.
Phenol is an important commodity chemical, and is a starting material for the production of numerous industrial chemicals and polymers, including bisphenol A and phenolic resins, and others. At present, the production of phenol entirely depends on the chemical synthesis from benzene, and its annual production exceeds 8 million tons worldwide. Microbial production of phenol seems to be a non-viable process considering the high toxicity of phenol to the cell.
In the paper published online in Biotechnology Journal, a Korean research team led by Distinguished Professor Sang Yup Lee at the Department of Chemical and Biomolecular Engineering from the Korea Advanced Institute of Science and Technology (KAIST) reported the successful development of an engineered Escherichia coli (E. coli) strain which can produce phenol from glucose. E. coli has been a workhorse for biological production of various value-added compounds such as succinic acid and 1,4-butanediol in industrial scale. However, due to its low tolerance to phenol, E. coli was not considered a viable host strain for the biological production of phenol.
Professor Lee"s team, a leading research group in metabolic engineering, noted the genetic and physiological differences of various E. coli strains and investigated 18 different E. coli strains with respect to phenol tolerance and engineered all of the 18 strains simultaneously. If the traditional genetic engineering methods were used, this work would have taken years to do. To overcome this challenge, the research team used synthetic small RNA (sRNA) technology they recently developed (Nature Biotechnology, vol 31, pp 170-174, 2013). The sRNA technology allowed the team to screen 18 E. coli strains with respect to the phenol tolerance, and the activities of the metabolic pathway and enzyme involved in the production of phenol. The research team also metabolically engineered the E. coli strains to increase carbon flux toward phenol and finally generated an engineered E. coli strain which can produce phenol from glucose.
Furthermore, the team developed a biphasic extractive fermentation process to minimize the toxicity of phenol to E. coli cells. Glycerol tributyrate was found to have low toxicity to E. coli and allowed efficient extraction of phenol from the culture broth. Through the biphasic fed-batch fermentation using glycerol tributyrate as an in situ extractant, the final engineered E. coli strain produced phenol to the highest titer and productivity reported (3.8 g/L and 0.18 g/L/h, respectively). The strategy used for the strain development and the fermentation process will serve as a framework for metabolic engineering of microorganisms for the production of toxic chemicals from renewable resources.
This work was supported by the Intelligent Synthetic Biology Center through the Global Frontier Project (2011-0031963) of the Ministry of Science, ICT & Future Planning through the National Research Foundation of Korea.
Process of Phenol Production
2013.11.05 View 8074 -
 KAIST announced a novel technology to produce gasoline by a metabolically engineered microorganism
A major scientific breakthrough in the development of renewable energy sources and other important chemicals; The research team succeeded in producing 580 mg of gasoline per liter of cultured broth by converting in vivo generated fatty acids
For many decades, we have been relying on fossil resources to produce liquid fuels such as gasoline, diesel, and many industrial and consumer chemicals for daily use. However, increasing strains on natural resources as well as environmental issues including global warming have triggered a strong interest in developing sustainable ways to obtain fuels and chemicals.
Gasoline, the petroleum-derived product that is most widely used as a fuel for transportation, is a mixture of hydrocarbons, additives, and blending agents. The hydrocarbons, called alkanes, consist only of carbon and hydrogen atoms. Gasoline has a combination of straight-chain and branched-chain alkanes (hydrocarbons) consisted of 4-12 carbon atoms linked by direct carbon-carbon bonds.
Previously, through metabolic engineering of Escherichia coli (E. coli), there have been a few research results on the production of long-chain alkanes, which consist of 13-17 carbon atoms, suitable for replacing diesel. However, there has been no report on the microbial production of short-chain alkanes, a possible substitute for gasoline.
In the paper (entitled "Microbial Production of Short-chain Alkanes") published online in Nature on September 29, a Korean research team led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at the Korea Advanced Institute of Science and Technology (KAIST) reported, for the first time, the development of a novel strategy for microbial gasoline production through metabolic engineering of E. coli.
The research team engineered the fatty acid metabolism to provide the fatty acid derivatives that are shorter than normal intracellular fatty acid metabolites, and introduced a novel synthetic pathway for the biosynthesis of short-chain alkanes. This allowed the development of platform E. coli strain capable of producing gasoline for the first time. Furthermore, this platform strain, if desired, can be modified to produce other products such as short-chain fatty esters and short-chain fatty alcohols.
In this paper, the Korean researchers described detailed strategies for 1) screening of enzymes associated with the production of fatty acids, 2) engineering of enzymes and fatty acid biosynthetic pathways to concentrate carbon flux towards the short-chain fatty acid production, and 3) converting short-chain fatty acids to their corresponding alkanes (gasoline) by introducing a novel synthetic pathway and optimization of culture conditions. Furthermore, the research team showed the possibility of producing fatty esters and alcohols by introducing responsible enzymes into the same platform strain.
Professor Sang Yup Lee said, "It is only the beginning of the work towards sustainable production of gasoline. The titer is rather low due to the low metabolic flux towards the formation of short-chain fatty acids and their derivatives. We are currently working on increasing the titer, yield and productivity of bio-gasoline. Nonetheless, we are pleased to report, for the first time, the production of gasoline through the metabolic engineering of E. coli, which we hope will serve as a basis for the metabolic engineering of microorganisms to produce fuels and chemicals from renewable resources."
This research was supported by the Advanced Biomass Research and Development Center of Korea (ABC-2010-0029799) through the Global Frontier Research Program of the Ministry of Science, ICT and Future Planning (MSIP) through the National Research Foundation (NRF), Republic of Korea. Systems metabolic engineering work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012-C1AAA001-2012M1A2A2026556) by MSIP through NRF.
Short-Chain Alkanes Generated from Renewable Biomass
This diagram shows the metabolic engineering of Escherichia coli for the production of short-chain alkanes (gasoline) from renewable biomass.
Nature Cover Page (September 29th, 2013)
2013.11.04 View 9280
KAIST announced a novel technology to produce gasoline by a metabolically engineered microorganism
A major scientific breakthrough in the development of renewable energy sources and other important chemicals; The research team succeeded in producing 580 mg of gasoline per liter of cultured broth by converting in vivo generated fatty acids
For many decades, we have been relying on fossil resources to produce liquid fuels such as gasoline, diesel, and many industrial and consumer chemicals for daily use. However, increasing strains on natural resources as well as environmental issues including global warming have triggered a strong interest in developing sustainable ways to obtain fuels and chemicals.
Gasoline, the petroleum-derived product that is most widely used as a fuel for transportation, is a mixture of hydrocarbons, additives, and blending agents. The hydrocarbons, called alkanes, consist only of carbon and hydrogen atoms. Gasoline has a combination of straight-chain and branched-chain alkanes (hydrocarbons) consisted of 4-12 carbon atoms linked by direct carbon-carbon bonds.
Previously, through metabolic engineering of Escherichia coli (E. coli), there have been a few research results on the production of long-chain alkanes, which consist of 13-17 carbon atoms, suitable for replacing diesel. However, there has been no report on the microbial production of short-chain alkanes, a possible substitute for gasoline.
In the paper (entitled "Microbial Production of Short-chain Alkanes") published online in Nature on September 29, a Korean research team led by Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at the Korea Advanced Institute of Science and Technology (KAIST) reported, for the first time, the development of a novel strategy for microbial gasoline production through metabolic engineering of E. coli.
The research team engineered the fatty acid metabolism to provide the fatty acid derivatives that are shorter than normal intracellular fatty acid metabolites, and introduced a novel synthetic pathway for the biosynthesis of short-chain alkanes. This allowed the development of platform E. coli strain capable of producing gasoline for the first time. Furthermore, this platform strain, if desired, can be modified to produce other products such as short-chain fatty esters and short-chain fatty alcohols.
In this paper, the Korean researchers described detailed strategies for 1) screening of enzymes associated with the production of fatty acids, 2) engineering of enzymes and fatty acid biosynthetic pathways to concentrate carbon flux towards the short-chain fatty acid production, and 3) converting short-chain fatty acids to their corresponding alkanes (gasoline) by introducing a novel synthetic pathway and optimization of culture conditions. Furthermore, the research team showed the possibility of producing fatty esters and alcohols by introducing responsible enzymes into the same platform strain.
Professor Sang Yup Lee said, "It is only the beginning of the work towards sustainable production of gasoline. The titer is rather low due to the low metabolic flux towards the formation of short-chain fatty acids and their derivatives. We are currently working on increasing the titer, yield and productivity of bio-gasoline. Nonetheless, we are pleased to report, for the first time, the production of gasoline through the metabolic engineering of E. coli, which we hope will serve as a basis for the metabolic engineering of microorganisms to produce fuels and chemicals from renewable resources."
This research was supported by the Advanced Biomass Research and Development Center of Korea (ABC-2010-0029799) through the Global Frontier Research Program of the Ministry of Science, ICT and Future Planning (MSIP) through the National Research Foundation (NRF), Republic of Korea. Systems metabolic engineering work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012-C1AAA001-2012M1A2A2026556) by MSIP through NRF.
Short-Chain Alkanes Generated from Renewable Biomass
This diagram shows the metabolic engineering of Escherichia coli for the production of short-chain alkanes (gasoline) from renewable biomass.
Nature Cover Page (September 29th, 2013)
2013.11.04 View 9280 -
 A powerful strategy for developing microbial cell factories by employing synthetic small RNAs
The current systems for the production of chemicals, fuels and materials heavily rely on the use of fossil resources. Due to the increasing concerns on climate change and other environmental problems, however, there has been much interest in developing biorefineries for the production of such chemicals, fuels and materials from renewable resources. For the biorefineries to be competitive with the traditional fossil resource-based refineries, development of high performance microorganisms is the most important as it will affect the overall economics of the process most significantly. Metabolic engineering, which can be defined as purposeful modification of cellular metabolic and regulatory networks with an aim to improve the production of a desired product, has been successfully employed to improve the performance of the cell. However, it is not trivial to engineer the cellular metabolism and regulatory circuits in the cell due to their high complexity.
In metabolic engineering, it is important to find the genes that need to be amplified and attenuated in order to increase the product formation rate while minimizing the production of undesirable byproducts. Gene knock-out experiments are often performed to delete those metabolic fluxes that will consequently result in the increase of the desired product formation. However, gene knock-out experiments require much effort and time to perform, and are difficult to do for a large number of genes. Furthermore, the gene knock-out experiments performed in one strain cannot be transferred to another organism and thus the whole experimental process has to be repeated. This is a big problem in developing a high performance microbial cell factory because it is required to find the best platform strain among many different strains. Therefore, researchers have been eager to develop a strategy that allows rapid identification of multiple genes to be attenuated in multiple strains at the same time.
A Korean research team led by Distinguished Professor Sang Yup Lee at the Department of Chemical and Biomolecular Engineering from the Korea Advanced Institute of Science and Technology (KAIST) reported the development of a strategy for efficiently developing microbial cell factories by employing synthetic small RNAs (sRNAs). They first reported the development of such system in Nature Biotechnology last February. This strategy of employing synthetic sRNAs in metabolic engineering has been receiving great interest worldwide as it allows easy, rapid, high-throughput, tunable, and un-doable knock-down of multiple genes in multiple strains at the same time. The research team published a paper online on August 8 as a cover page (September issue) in Nature Protocols, describing the detailed strategy and protocol to employ synthetic sRNAs for metabolic engineering.
In this paper, researchers described the detailed step-by-step protocol for synthetic sRNA-based gene expression control, including the sRNA design principles. Tailor-made synthetic sRNAs can be easily manipulated by using conventional gene cloning method. The use of synthetic sRNAs for gene expression regulation provides several advantages such as portability, conditionality, and tunability in high-throughput experiments. Plasmid-based synthetic sRNA expression system does not leave any scar on the chromosome, and can be easily transferred to many other host strains to be examined. Thus, the construction of libraries and examination of different host strains are much easier than the conventional hard-coded gene manipulation systems. Also, the expression of genes can be conditionally repressed by controlling the production of synthetic sRNAs. Synthetic sRNAs possessing different repression efficiencies make it possible to finely tune the gene expression levels as well. Furthermore, synthetic sRNAs allow knock-down of the expression of essential genes, which was not possible by conventional gene knock-out experiments.
Synthetic sRNAs can be utilized for diverse experiments where gene expression regulation is needed. One of promising applications is high-throughput screening of the target genes to be manipulated and multiple strains simultaneously to enhance the production of chemicals and materials of interest. Such simultaneous optimization of gene targets and strains has been one of the big challenges in metabolic engineering. Another application is to fine tune the expression of the screened genes for flux optimization, which would enhance chemical production further by balancing the flux between biomass formation and target chemical production. Synthetic sRNAs can also be applied to finely regulating genetic interactions in a circuit or network, which is essential in synthetic biology. Once a sRNA scaffold-harboring plasmid is constructed, tailor-made, synthetic sRNAs can be made within 3-4 days, followed by the desired application experiments.
Dr. Eytan Zlotorynski, an editor at Nature Protocols, said "This paper describes the detailed protocol for the design and applications of synthetic sRNA. The method, which has many advantages, is likely to become common practice, and prove useful for metabolic engineering and synthetic biology studies."
This paper published in Nature Protocols will be useful for all researchers in academia and industry who are interested in the use of synthetic sRNAs for fundamental and applied biological and biotechnological studies.
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012-C1AAA001-2012M1A2A2026556) and the Intelligent Synthetic Biology Center through the Global Frontier Project (2011-0031963) of the Ministry of Science, ICT and Future Planning through the National Research Foundation of Korea.
2013.10.31 View 7245
A powerful strategy for developing microbial cell factories by employing synthetic small RNAs
The current systems for the production of chemicals, fuels and materials heavily rely on the use of fossil resources. Due to the increasing concerns on climate change and other environmental problems, however, there has been much interest in developing biorefineries for the production of such chemicals, fuels and materials from renewable resources. For the biorefineries to be competitive with the traditional fossil resource-based refineries, development of high performance microorganisms is the most important as it will affect the overall economics of the process most significantly. Metabolic engineering, which can be defined as purposeful modification of cellular metabolic and regulatory networks with an aim to improve the production of a desired product, has been successfully employed to improve the performance of the cell. However, it is not trivial to engineer the cellular metabolism and regulatory circuits in the cell due to their high complexity.
In metabolic engineering, it is important to find the genes that need to be amplified and attenuated in order to increase the product formation rate while minimizing the production of undesirable byproducts. Gene knock-out experiments are often performed to delete those metabolic fluxes that will consequently result in the increase of the desired product formation. However, gene knock-out experiments require much effort and time to perform, and are difficult to do for a large number of genes. Furthermore, the gene knock-out experiments performed in one strain cannot be transferred to another organism and thus the whole experimental process has to be repeated. This is a big problem in developing a high performance microbial cell factory because it is required to find the best platform strain among many different strains. Therefore, researchers have been eager to develop a strategy that allows rapid identification of multiple genes to be attenuated in multiple strains at the same time.
A Korean research team led by Distinguished Professor Sang Yup Lee at the Department of Chemical and Biomolecular Engineering from the Korea Advanced Institute of Science and Technology (KAIST) reported the development of a strategy for efficiently developing microbial cell factories by employing synthetic small RNAs (sRNAs). They first reported the development of such system in Nature Biotechnology last February. This strategy of employing synthetic sRNAs in metabolic engineering has been receiving great interest worldwide as it allows easy, rapid, high-throughput, tunable, and un-doable knock-down of multiple genes in multiple strains at the same time. The research team published a paper online on August 8 as a cover page (September issue) in Nature Protocols, describing the detailed strategy and protocol to employ synthetic sRNAs for metabolic engineering.
In this paper, researchers described the detailed step-by-step protocol for synthetic sRNA-based gene expression control, including the sRNA design principles. Tailor-made synthetic sRNAs can be easily manipulated by using conventional gene cloning method. The use of synthetic sRNAs for gene expression regulation provides several advantages such as portability, conditionality, and tunability in high-throughput experiments. Plasmid-based synthetic sRNA expression system does not leave any scar on the chromosome, and can be easily transferred to many other host strains to be examined. Thus, the construction of libraries and examination of different host strains are much easier than the conventional hard-coded gene manipulation systems. Also, the expression of genes can be conditionally repressed by controlling the production of synthetic sRNAs. Synthetic sRNAs possessing different repression efficiencies make it possible to finely tune the gene expression levels as well. Furthermore, synthetic sRNAs allow knock-down of the expression of essential genes, which was not possible by conventional gene knock-out experiments.
Synthetic sRNAs can be utilized for diverse experiments where gene expression regulation is needed. One of promising applications is high-throughput screening of the target genes to be manipulated and multiple strains simultaneously to enhance the production of chemicals and materials of interest. Such simultaneous optimization of gene targets and strains has been one of the big challenges in metabolic engineering. Another application is to fine tune the expression of the screened genes for flux optimization, which would enhance chemical production further by balancing the flux between biomass formation and target chemical production. Synthetic sRNAs can also be applied to finely regulating genetic interactions in a circuit or network, which is essential in synthetic biology. Once a sRNA scaffold-harboring plasmid is constructed, tailor-made, synthetic sRNAs can be made within 3-4 days, followed by the desired application experiments.
Dr. Eytan Zlotorynski, an editor at Nature Protocols, said "This paper describes the detailed protocol for the design and applications of synthetic sRNA. The method, which has many advantages, is likely to become common practice, and prove useful for metabolic engineering and synthetic biology studies."
This paper published in Nature Protocols will be useful for all researchers in academia and industry who are interested in the use of synthetic sRNAs for fundamental and applied biological and biotechnological studies.
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012-C1AAA001-2012M1A2A2026556) and the Intelligent Synthetic Biology Center through the Global Frontier Project (2011-0031963) of the Ministry of Science, ICT and Future Planning through the National Research Foundation of Korea.
2013.10.31 View 7245 -
 Distinguished Professor Sang Yup Lee appointed as an advisor for Shanghai Jiao Tong University in China
In recognition of his outstanding accomplishments in the area of bioengineering, specializing in metabolic engineering, Sang Yup Lee, a distinguished professor of Chemical & Biomolecular Engineering at KAIST, was assigned as an advisory professor for the bioengineering department at Shanghai Jiao Tong University in China for five years from August 2013 to July 2018.
Together with Peking University and Tsinghua University, Shanghai Jiao Tong University is one of the top three universities in China.
The advisory professors carry out collaborated research programs in special areas and provide advice on education and research issues.
Professor Lee, a specialist in metabolic engineering, has initiated systems metabolic engineering which integrates metabolic engineering, systems biology, and synthetic biology and has applied it to various chemical production systems to develop bio fuel and many eco-friendly chemical production processes. Recently, he received the Marvin J. Johnson Award from the American Chemistry Society, the Charles Thom Award from the American Society for Industrial Microbiology, as well as the Amgen Biochemical Engineering Award. As a global leader in the area of bioengineering, Professor Lee is a member of the Korean Academy of Science & Technology, the National Academy of Engineering of Korea, the US National Academy of Engineering, and is the chairman of the Global Agenda Council on Biotechnology at the World Economic Forum.
2013.10.31 View 6908
Distinguished Professor Sang Yup Lee appointed as an advisor for Shanghai Jiao Tong University in China
In recognition of his outstanding accomplishments in the area of bioengineering, specializing in metabolic engineering, Sang Yup Lee, a distinguished professor of Chemical & Biomolecular Engineering at KAIST, was assigned as an advisory professor for the bioengineering department at Shanghai Jiao Tong University in China for five years from August 2013 to July 2018.
Together with Peking University and Tsinghua University, Shanghai Jiao Tong University is one of the top three universities in China.
The advisory professors carry out collaborated research programs in special areas and provide advice on education and research issues.
Professor Lee, a specialist in metabolic engineering, has initiated systems metabolic engineering which integrates metabolic engineering, systems biology, and synthetic biology and has applied it to various chemical production systems to develop bio fuel and many eco-friendly chemical production processes. Recently, he received the Marvin J. Johnson Award from the American Chemistry Society, the Charles Thom Award from the American Society for Industrial Microbiology, as well as the Amgen Biochemical Engineering Award. As a global leader in the area of bioengineering, Professor Lee is a member of the Korean Academy of Science & Technology, the National Academy of Engineering of Korea, the US National Academy of Engineering, and is the chairman of the Global Agenda Council on Biotechnology at the World Economic Forum.
2013.10.31 View 6908 -
 Top Ten Ways Biotechnology Could Improve Our Everyday Life
The Global Agenda Council on Biotechnology, one of the global networks under the World Economic Forum, which is composed of the world’s leading experts in the field of biotechnology, announced on February 25, 2013 that the council has indentified “ten most important biotechnologies” that could help meet rapidly growing demand for energy, food, nutrition, and health. These new technologies, the council said, also have the potential to increase productivity and create new jobs.
“The technologies selected by the members of the Global Agenda Council on Biotechnology represent almost all types of biotechnology.Utilization of waste, personalized medicine,and ocean agricultureare examples of the challenges where biotechnology can offer solutions,”said Sang Yup Lee, Chair of the Global Agenda Council on Biotechnology and Distinguished Professor in the Department of Chemical and Biomolecular Engineering at the Korea Advanced Institute of Science and Technology (KAIST). He also added that “the members of the council concluded that regulatory certainty, public perception, and investment are the key enablers for the growth of biotechnology.”
These ideas will be further explored during “Biotechnology Week” at the World Economic Forum’s Blog (http://wef.ch/blog) from Monday, 25 February, 2013. The full list follows below:
Bio-based sustainable production of chemicals, energy, fuels and materials
Through the last century, human activity has depleted approximately half of the world’s reserves of fossil hydrocarbons. These reserves, which took over 600 million years to accumulate, are non-renewable and their extraction, refining and use contribute significantly to human emissions of greenhouse gases and the warming of our planet. In order to sustain human development going forward, a carbon-neutral alternative must be implemented. The key promising technology is biological synthesis; that is, bio-based production of chemicals, fuels and materials from plants that can be re-grown.
Engineering sustainable food production
The continuing increase in our numbers and affluence are posing growing challenges to the ability of humanity to produce adequate food (as well as feed, and now fuel). Although controversial, modern genetic modification of crops has supported growth in agricultural productivity. In 2011, 16.7 million farmers grew biotechnology-developed crops on almost 400 million acres in 29 countries, 19 of which were developing countries. Properly managed, such crops have the potential to lower both pesticide use and tilling which erodes soil.
Sea-water based bio-processes
Over 70% of the earth surface is covered by seawater, and it is the most abundant water source available on the planet. But we are yet to discover the full potential of it. For example with halliophic bacteria capable of growing in the seawater can be engineered to grow faster and produce useful products including chemicals, fuels and polymeric materials. Ocean agriculture is also a promising technology. It is based on the photosynthetic biomass from the oceans, like macroalgae and microalgae.
Non-resource draining zero waste bio-processing
The sustainable goal of zero waste may become a reality with biotechnology. Waste streams can be processed at bio-refineries and turned into valuable chemicals and fuels, thereby closing the loop of production with no net waste. Advances in biotechnology are now allowing lower cost, less draining inputs to be used, including methane, and waste heat. These advances are simplifying waste streams with the potential to reduce toxicity as well as support their use in other processes, moving society progressively closer to the sustainable goal of zero waste.
Using carbon dioxide as a raw material
Biotechnology is poised to contribute solutions to mitigate the growing threat of rising CO2 levels. Recent advances are rapidly increasing our understanding of how living organisms consume and use CO2. By harnessing the power of these natural biological systems, scientists are engineering a new wave of approaches to convert waste CO2 and C1 molecules into energy, fuels, chemicals, and new materials.
Regenerative medicine
Regenerative medicine has become increasingly important due to both increased longevity and treatment of injury. Tissue engineering based on various bio-materials has been developed to speed up the regenerative medicine. Recently, stem cells, especially the induced pluripotent stem cells (iPS), have provided another great opportunity for regenerative medicine. Combination of tissue engineering and stem cell (including iPS) technologies will allow replacements of damaged or old human organs with functional ones in the near future.
Rapid and precise development and manufacturing of medicine and vaccines
A global pandemic remains one of the most real and serious threats to humanity. Biotechnology has the potential to rapidly identify biological threats, develop and manufacture potential cures. Leading edge biotechnology is now offering the potential to rapidly produce therapeutics and vaccines against virtually any target. These technologies, including messenger therapeutics, targeted immunotherapies, conjugated nanoparticles, and structure-based engineering, have already produced candidates with substantial potential to improve human health globally.
Accurate, fast, cheap, and personalized diagnostics and prognostics
Identification of better targets and combining nanotechnology and information technology it will be possible to develop rapid, accurate, personalized and inexpensive diagnostics and prognostics systems.
Bio-tech improvements to soil and water
Arable land and fresh water are two of the most important, yet limited, resources on earth. Abuse and mis-appropriation have threatened these resources, as the demand on them has increased. Advances in biotechnology have already yielded technologies that can restore the vitality and viability of these resources. A new generation of technologies: bio-remediation, bio-regeneration and bio-augmentation are being developed, offering the potential to not only further restore these resources, but also augment their potential.
Advanced healthcare through genome sequencing
It took more than 13 years and $1.5 billion to sequence the first human genome and today we can sequence a complete human genome in a single day for less than $1,000. When we analyze the roughly 3 billion base pairs in such a sequence we find that we differ from each other in several million of these base pairs. In the vast majority of cases these difference do not cause any issues but in rare cases they cause disease, or susceptibility to disease. Medical research and practice will increasingly be driven by our understanding of such genetic variations together with their phenotypic consequences.
2013.03.19 View 9348
Top Ten Ways Biotechnology Could Improve Our Everyday Life
The Global Agenda Council on Biotechnology, one of the global networks under the World Economic Forum, which is composed of the world’s leading experts in the field of biotechnology, announced on February 25, 2013 that the council has indentified “ten most important biotechnologies” that could help meet rapidly growing demand for energy, food, nutrition, and health. These new technologies, the council said, also have the potential to increase productivity and create new jobs.
“The technologies selected by the members of the Global Agenda Council on Biotechnology represent almost all types of biotechnology.Utilization of waste, personalized medicine,and ocean agricultureare examples of the challenges where biotechnology can offer solutions,”said Sang Yup Lee, Chair of the Global Agenda Council on Biotechnology and Distinguished Professor in the Department of Chemical and Biomolecular Engineering at the Korea Advanced Institute of Science and Technology (KAIST). He also added that “the members of the council concluded that regulatory certainty, public perception, and investment are the key enablers for the growth of biotechnology.”
These ideas will be further explored during “Biotechnology Week” at the World Economic Forum’s Blog (http://wef.ch/blog) from Monday, 25 February, 2013. The full list follows below:
Bio-based sustainable production of chemicals, energy, fuels and materials
Through the last century, human activity has depleted approximately half of the world’s reserves of fossil hydrocarbons. These reserves, which took over 600 million years to accumulate, are non-renewable and their extraction, refining and use contribute significantly to human emissions of greenhouse gases and the warming of our planet. In order to sustain human development going forward, a carbon-neutral alternative must be implemented. The key promising technology is biological synthesis; that is, bio-based production of chemicals, fuels and materials from plants that can be re-grown.
Engineering sustainable food production
The continuing increase in our numbers and affluence are posing growing challenges to the ability of humanity to produce adequate food (as well as feed, and now fuel). Although controversial, modern genetic modification of crops has supported growth in agricultural productivity. In 2011, 16.7 million farmers grew biotechnology-developed crops on almost 400 million acres in 29 countries, 19 of which were developing countries. Properly managed, such crops have the potential to lower both pesticide use and tilling which erodes soil.
Sea-water based bio-processes
Over 70% of the earth surface is covered by seawater, and it is the most abundant water source available on the planet. But we are yet to discover the full potential of it. For example with halliophic bacteria capable of growing in the seawater can be engineered to grow faster and produce useful products including chemicals, fuels and polymeric materials. Ocean agriculture is also a promising technology. It is based on the photosynthetic biomass from the oceans, like macroalgae and microalgae.
Non-resource draining zero waste bio-processing
The sustainable goal of zero waste may become a reality with biotechnology. Waste streams can be processed at bio-refineries and turned into valuable chemicals and fuels, thereby closing the loop of production with no net waste. Advances in biotechnology are now allowing lower cost, less draining inputs to be used, including methane, and waste heat. These advances are simplifying waste streams with the potential to reduce toxicity as well as support their use in other processes, moving society progressively closer to the sustainable goal of zero waste.
Using carbon dioxide as a raw material
Biotechnology is poised to contribute solutions to mitigate the growing threat of rising CO2 levels. Recent advances are rapidly increasing our understanding of how living organisms consume and use CO2. By harnessing the power of these natural biological systems, scientists are engineering a new wave of approaches to convert waste CO2 and C1 molecules into energy, fuels, chemicals, and new materials.
Regenerative medicine
Regenerative medicine has become increasingly important due to both increased longevity and treatment of injury. Tissue engineering based on various bio-materials has been developed to speed up the regenerative medicine. Recently, stem cells, especially the induced pluripotent stem cells (iPS), have provided another great opportunity for regenerative medicine. Combination of tissue engineering and stem cell (including iPS) technologies will allow replacements of damaged or old human organs with functional ones in the near future.
Rapid and precise development and manufacturing of medicine and vaccines
A global pandemic remains one of the most real and serious threats to humanity. Biotechnology has the potential to rapidly identify biological threats, develop and manufacture potential cures. Leading edge biotechnology is now offering the potential to rapidly produce therapeutics and vaccines against virtually any target. These technologies, including messenger therapeutics, targeted immunotherapies, conjugated nanoparticles, and structure-based engineering, have already produced candidates with substantial potential to improve human health globally.
Accurate, fast, cheap, and personalized diagnostics and prognostics
Identification of better targets and combining nanotechnology and information technology it will be possible to develop rapid, accurate, personalized and inexpensive diagnostics and prognostics systems.
Bio-tech improvements to soil and water
Arable land and fresh water are two of the most important, yet limited, resources on earth. Abuse and mis-appropriation have threatened these resources, as the demand on them has increased. Advances in biotechnology have already yielded technologies that can restore the vitality and viability of these resources. A new generation of technologies: bio-remediation, bio-regeneration and bio-augmentation are being developed, offering the potential to not only further restore these resources, but also augment their potential.
Advanced healthcare through genome sequencing
It took more than 13 years and $1.5 billion to sequence the first human genome and today we can sequence a complete human genome in a single day for less than $1,000. When we analyze the roughly 3 billion base pairs in such a sequence we find that we differ from each other in several million of these base pairs. In the vast majority of cases these difference do not cause any issues but in rare cases they cause disease, or susceptibility to disease. Medical research and practice will increasingly be driven by our understanding of such genetic variations together with their phenotypic consequences.
2013.03.19 View 9348 -
 An efficient strategy for developing microbial cell factories by employing synthetic small regulatory RNAs
A new metabolic engineering tool that allows fine control of gene expression level by employing synthetic small regulatory RNAs was developed to efficiently construct microbial cell factories producing desired chemicals and materials
Biotechnologists have been working hard to address the climate change and limited fossil resource issues through the development of sustainable processes for the production of chemicals, fuels and materials from renewable non-food biomass. One promising sustainable technology is the use of microbial cell factories for the efficient production of desired chemicals and materials. When microorganisms are isolated from nature, the performance in producing our desired product is rather poor. That is why metabolic engineering is performed to improve the metabolic and cellular characteristics to achieve enhanced production of desired product at high yield and productivity. Since the performance of microbial cell factory is very important in lowering the overall production cost of the bioprocess, many different strategies and tools have been developed for the metabolic engineering of microorganisms.
One of the big challenges in metabolic engineering is to find the best platform organism and to find those genes to be engineered so as to maximize the production efficiency of the desired chemical. Even Escherichia coli, the most widely utilized simple microorganism, has thousands of genes, the expression of which is highly regulated and interconnected to finely control cellular and metabolic activities. Thus, the complexity of cellular genetic interactions is beyond our intuition and thus it is very difficult to find effective target genes to engineer. Together with gene amplification strategy, gene knockout strategy has been an essential tool in metabolic engineering to redirect the pathway fluxes toward our desired product formation. However, experiment to engineer many genes can be rather difficult due to the time and effort required; for example, gene deletion experiment can take a few weeks depending on the microorganisms. Furthermore, as certain genes are essential or play important roles for the survival of a microorganism, gene knockout experiments cannot be performed. Even worse, there are many different microbial strains one can employ. There are more than 50 different E. coli strains that metabolic engineer can consider to use. Since gene knockout experiment is hard-coded (that is, one should repeat the gene knockout experiments for each strain), the result cannot be easily transferred from one strain to another.
A paper published in Nature Biotechnology online today addresses this issue and suggests a new strategy for identifying gene targets to be knocked out or knocked down through the use of synthetic small RNA. A Korean research team led by Distinguished Professor Sang Yup Lee at the Department of Chemical and Biomolecular Engineering, Korea Advanced Institute of Science and Technology (KAIST), a prestigeous science and engineering university in Korea reported that synthetic small RNA can be employed for finely controlling the expression levels of multiple genes at the translation level. Already well-known for their systems metabolic engineering strategies, Professor Lee’s team added one more strategy to efficiently develop microbial cell factories for the production of chemicals and materials.
Gene expression works like this: the hard-coded blueprint (DNA) is transcribed into messenger RNA (mRNA), and the coding information in mRNA is read to produce protein by ribosomes. Conventional genetic engineering approaches have often targeted modification of the blueprint itself (DNA) to alter organism’s physiological characteristics. Again, engineering the blueprint itself takes much time and effort, and in addition, the results obtained cannot be transferred to another organism without repeating the whole set of experiments. This is why Professor Lee and his colleagues aimed at controlling the gene expression level at the translation stage through the use of synthetic small RNA. They created novel RNAs that can regulate the translation of multiple messenger RNAs (mRNA), and consequently varying the expression levels of multiple genes at the same time. Briefly, synthetic regulatory RNAs interrupt gene expression process from DNA to protein by destroying the messenger RNAs to different yet controllable extents. The advantages of taking this strategy of employing synthetic small regulatory RNAs include simple, easy and high-throughput identification of gene knockout or knockdown targets, fine control of gene expression levels, transferability to many different host strains, and possibility of identifying those gene targets that are essential.
As proof-of-concept demonstration of the usefulness of this strategy, Professor Lee and his colleagues applied it to develop engineered E. coli strains capable of producing an aromatic amino acid tyrosine, which is used for stress symptom relief, food supplements, and precursor for many drugs. They examined a large number of genes in multiple E. coli strains, and developed a highly efficient tyrosine producer. Also, they were able to show that this strategy can be employed to an already metabolically engineered E. coli strain for further improvement by demonstrating the development of highly efficient producer of cadaverine, an important platform chemical for nylon in the chemical industry.
This new strategy, being simple yet very powerful for systems metabolic engineering, is thus expected to facilitate the efficient development of microbial cell factories capable of producing chemicals, fuels and materials from renewable biomass.
Source: Dokyun Na, Seung Min Yoo, Hannah Chung, Hyegwon Park, Jin Hwan Park, and Sang Yup Lee, “Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs”, Nature Biotechnology, doi:10.1038/nbt.2461 (2013)
2013.03.19 View 7978
An efficient strategy for developing microbial cell factories by employing synthetic small regulatory RNAs
A new metabolic engineering tool that allows fine control of gene expression level by employing synthetic small regulatory RNAs was developed to efficiently construct microbial cell factories producing desired chemicals and materials
Biotechnologists have been working hard to address the climate change and limited fossil resource issues through the development of sustainable processes for the production of chemicals, fuels and materials from renewable non-food biomass. One promising sustainable technology is the use of microbial cell factories for the efficient production of desired chemicals and materials. When microorganisms are isolated from nature, the performance in producing our desired product is rather poor. That is why metabolic engineering is performed to improve the metabolic and cellular characteristics to achieve enhanced production of desired product at high yield and productivity. Since the performance of microbial cell factory is very important in lowering the overall production cost of the bioprocess, many different strategies and tools have been developed for the metabolic engineering of microorganisms.
One of the big challenges in metabolic engineering is to find the best platform organism and to find those genes to be engineered so as to maximize the production efficiency of the desired chemical. Even Escherichia coli, the most widely utilized simple microorganism, has thousands of genes, the expression of which is highly regulated and interconnected to finely control cellular and metabolic activities. Thus, the complexity of cellular genetic interactions is beyond our intuition and thus it is very difficult to find effective target genes to engineer. Together with gene amplification strategy, gene knockout strategy has been an essential tool in metabolic engineering to redirect the pathway fluxes toward our desired product formation. However, experiment to engineer many genes can be rather difficult due to the time and effort required; for example, gene deletion experiment can take a few weeks depending on the microorganisms. Furthermore, as certain genes are essential or play important roles for the survival of a microorganism, gene knockout experiments cannot be performed. Even worse, there are many different microbial strains one can employ. There are more than 50 different E. coli strains that metabolic engineer can consider to use. Since gene knockout experiment is hard-coded (that is, one should repeat the gene knockout experiments for each strain), the result cannot be easily transferred from one strain to another.
A paper published in Nature Biotechnology online today addresses this issue and suggests a new strategy for identifying gene targets to be knocked out or knocked down through the use of synthetic small RNA. A Korean research team led by Distinguished Professor Sang Yup Lee at the Department of Chemical and Biomolecular Engineering, Korea Advanced Institute of Science and Technology (KAIST), a prestigeous science and engineering university in Korea reported that synthetic small RNA can be employed for finely controlling the expression levels of multiple genes at the translation level. Already well-known for their systems metabolic engineering strategies, Professor Lee’s team added one more strategy to efficiently develop microbial cell factories for the production of chemicals and materials.
Gene expression works like this: the hard-coded blueprint (DNA) is transcribed into messenger RNA (mRNA), and the coding information in mRNA is read to produce protein by ribosomes. Conventional genetic engineering approaches have often targeted modification of the blueprint itself (DNA) to alter organism’s physiological characteristics. Again, engineering the blueprint itself takes much time and effort, and in addition, the results obtained cannot be transferred to another organism without repeating the whole set of experiments. This is why Professor Lee and his colleagues aimed at controlling the gene expression level at the translation stage through the use of synthetic small RNA. They created novel RNAs that can regulate the translation of multiple messenger RNAs (mRNA), and consequently varying the expression levels of multiple genes at the same time. Briefly, synthetic regulatory RNAs interrupt gene expression process from DNA to protein by destroying the messenger RNAs to different yet controllable extents. The advantages of taking this strategy of employing synthetic small regulatory RNAs include simple, easy and high-throughput identification of gene knockout or knockdown targets, fine control of gene expression levels, transferability to many different host strains, and possibility of identifying those gene targets that are essential.
As proof-of-concept demonstration of the usefulness of this strategy, Professor Lee and his colleagues applied it to develop engineered E. coli strains capable of producing an aromatic amino acid tyrosine, which is used for stress symptom relief, food supplements, and precursor for many drugs. They examined a large number of genes in multiple E. coli strains, and developed a highly efficient tyrosine producer. Also, they were able to show that this strategy can be employed to an already metabolically engineered E. coli strain for further improvement by demonstrating the development of highly efficient producer of cadaverine, an important platform chemical for nylon in the chemical industry.
This new strategy, being simple yet very powerful for systems metabolic engineering, is thus expected to facilitate the efficient development of microbial cell factories capable of producing chemicals, fuels and materials from renewable biomass.
Source: Dokyun Na, Seung Min Yoo, Hannah Chung, Hyegwon Park, Jin Hwan Park, and Sang Yup Lee, “Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs”, Nature Biotechnology, doi:10.1038/nbt.2461 (2013)
2013.03.19 View 7978 -
 New BioFactory Technique Developed using sRNAs
Professor Sang Yup Lee
- published on the online edition of Nature Biotechnology. “Expected as a new strategy for the bio industry that may replace the chemical industry.”-
KAIST Chemical & Biomolecular engineering department’s Professor Sang Yup Lee and his team has developed a new technology that utilizes the synthetic small regulatory RNAs (sRNAs) to implement the BioFactory in a larger scale with more effectiveness.
* BioFactory: Microbial-based production system which creates the desired compound in mass by manipulating the genes of the cell.
In order to solve the problems of modern society, such as environmental pollution caused by the exhaustion of fossil fuels and usage of petrochemical products, an eco-friendly and sustainable bio industry is on the rise. BioFactory development technology has especially attracted the attention world-wide, with its ability to produce bio-energy, pharmaceuticals, eco-friendly materials and more.
For the development of an excellent BioFactory, selection for the gene that produces the desired compounds must be accompanied by finding the microorganism with high production efficiency; however, the previous research method had a complicated and time-consuming problem of having to manipulate the genes of the microorganism one by one.
Professor Sang Yup Lee’s research team, including Dr. Dokyun Na and Dr. Seung Min Yoo, has produced the synthetic sRNAs and utilized it to overcome the technical limitations mentioned above.
In particular, unlike the existing method, this technology using synthetic sRNAs exhibits no strain specificity which can dramatically shorten the experiment that used to take months to just a few days.
The research team applied the synthetic small regulatory RNA technology to the production of the tyrosine*, which is used as the precursor of the medicinal compound, and cadaverine**, widely utilized in a variety of petrochemical products, and has succeeded developing BioFactory with the world’s highest yield rate (21.9g /L, 12.6g / L each).
*tyrosine: amino acid known to control stress and improve concentration
**cadaverine: base material used in many petrochemical products, such as polyurethane
Professor Sang Yup Lee highlighted the significance of this research: “it is expected the synthetic small regulatory RNA technology will stimulate the BioFactory development and also serve as a catalyst which can make the chemical industry, currently represented by its petroleum energy, transform into bio industry.”
The study was carried out with the support of Global Frontier Project (Intelligent Bio-Systems Design and Synthesis Research Unit (Chief Seon Chang Kim)) and the findings have been published on January 20th in the online edition of the worldwide journal Nature Biotechnology.
2013.02.21 View 8458
New BioFactory Technique Developed using sRNAs
Professor Sang Yup Lee
- published on the online edition of Nature Biotechnology. “Expected as a new strategy for the bio industry that may replace the chemical industry.”-
KAIST Chemical & Biomolecular engineering department’s Professor Sang Yup Lee and his team has developed a new technology that utilizes the synthetic small regulatory RNAs (sRNAs) to implement the BioFactory in a larger scale with more effectiveness.
* BioFactory: Microbial-based production system which creates the desired compound in mass by manipulating the genes of the cell.
In order to solve the problems of modern society, such as environmental pollution caused by the exhaustion of fossil fuels and usage of petrochemical products, an eco-friendly and sustainable bio industry is on the rise. BioFactory development technology has especially attracted the attention world-wide, with its ability to produce bio-energy, pharmaceuticals, eco-friendly materials and more.
For the development of an excellent BioFactory, selection for the gene that produces the desired compounds must be accompanied by finding the microorganism with high production efficiency; however, the previous research method had a complicated and time-consuming problem of having to manipulate the genes of the microorganism one by one.
Professor Sang Yup Lee’s research team, including Dr. Dokyun Na and Dr. Seung Min Yoo, has produced the synthetic sRNAs and utilized it to overcome the technical limitations mentioned above.
In particular, unlike the existing method, this technology using synthetic sRNAs exhibits no strain specificity which can dramatically shorten the experiment that used to take months to just a few days.
The research team applied the synthetic small regulatory RNA technology to the production of the tyrosine*, which is used as the precursor of the medicinal compound, and cadaverine**, widely utilized in a variety of petrochemical products, and has succeeded developing BioFactory with the world’s highest yield rate (21.9g /L, 12.6g / L each).
*tyrosine: amino acid known to control stress and improve concentration
**cadaverine: base material used in many petrochemical products, such as polyurethane
Professor Sang Yup Lee highlighted the significance of this research: “it is expected the synthetic small regulatory RNA technology will stimulate the BioFactory development and also serve as a catalyst which can make the chemical industry, currently represented by its petroleum energy, transform into bio industry.”
The study was carried out with the support of Global Frontier Project (Intelligent Bio-Systems Design and Synthesis Research Unit (Chief Seon Chang Kim)) and the findings have been published on January 20th in the online edition of the worldwide journal Nature Biotechnology.
2013.02.21 View 8458 -
 Production of chemicals without petroleum
Systems metabolic engineering of microorganisms allows efficient production of natural and non-natural chemicals from renewable non-food biomass
In our everyday life, we use gasoline, diesel, plastics, rubbers, and numerous chemicals that are derived from fossil oil through petrochemical refinery processes. However, this is not sustainable due to the limited nature of fossil resources. Furthermore, our world is facing problems associated with climate change and other environmental problems due to the increasing use of fossil resources. One solution to address above problems is the use of renewable non-food biomass for the production of chemicals, fuels and materials through biorefineries. Microorganisms are used as biocatalysts for converting biomass to the products of interest. However, when microorganisms are isolated from nature, their efficiencies of producing our desired chemicals and materials are rather low. Metabolic engineering is thus performed to improve cellular characteristics to desired levels. Over the last decade, much advances have been made in systems biology that allows system-wide characterization of cellular networks, both qualitatively and quantitatively, followed by whole-cell level engineering based on these findings. Furthermore, rapid advances in synthetic biology allow design and synthesis of fine controlled metabolic and gene regulatory circuits. The strategies and methods of systems biology and synthetic biology are rapidly integrated with metabolic engineering, thus resulting in "systems metabolic engineering".
In the paper published online in Nature Chemical Biology on May 17, Professor Sang Yup Lee and his colleagues at the Department of Chemical and Biomolecular Engineering, Korea Advanced Institute of Science and Technology (KAIST), Daejeon, Korea present new general strategies of systems metabolic engineering for developing microorganisms for the production of natural and non-natural chemicals from renewable biomass. They first classified the chemicals to be produced into four categories based on whether they have thus far been identified to exist in nature (natural vs. nonnatural) and whether they can be produced by inherent pathways of microorganisms (inherent, noninherent, or created): natural-inherent, natural-noninherent, non-natural-noninherent, and non-natural-created ones. General strategies for systems metabolic engineering of microorganisms for the production of these chemicals using various tools and methods based on omics, genome-scale metabolic modeling and simulation, evolutionary engineering, synthetic biology are suggested with relevant examples. For the production of non-natural chemicals, strategies for the construction of synthetic metabolic pathways are also suggested. Having collected diverse tools and methods for systems metabolic engineering, authors also suggest how to use them and their possible limitations.
Professor Sang Yup Lee said "It is expected that increasing number of chemicals and materials will be produced through biorefineries. We are now equipped with new strategies for developing microbial strains that can produce our desired products at very high efficiencies, thus allowing cost competitiveness to those produced by petrochemical refineries."
Editor of Nature Chemical Biology, Dr. Catherine Goodman, said "It is exciting to see how quickly science is progressing in this field – ideas that used to be science fiction are taking shape in research labs and biorefineries. The article by Professor Lee and his colleagues not only highlights the most advanced techniques and strategies available, but offers critical advice to progress the field as a whole."
The works of Professor Lee have been supported by the Advanced Biomass Center and Intelligent Synthetic Biology Center of Global Frontier Program from the Korean Ministry of Education, Science and Technology through National Research Foundation.
Contact: Dr. Sang Yup Lee, Distinguished Professor and Dean, KAIST, Daejeon, Korea (leesy@kaist.ac.kr, +82-42-350-3930)
2012.05.23 View 9644
Production of chemicals without petroleum
Systems metabolic engineering of microorganisms allows efficient production of natural and non-natural chemicals from renewable non-food biomass
In our everyday life, we use gasoline, diesel, plastics, rubbers, and numerous chemicals that are derived from fossil oil through petrochemical refinery processes. However, this is not sustainable due to the limited nature of fossil resources. Furthermore, our world is facing problems associated with climate change and other environmental problems due to the increasing use of fossil resources. One solution to address above problems is the use of renewable non-food biomass for the production of chemicals, fuels and materials through biorefineries. Microorganisms are used as biocatalysts for converting biomass to the products of interest. However, when microorganisms are isolated from nature, their efficiencies of producing our desired chemicals and materials are rather low. Metabolic engineering is thus performed to improve cellular characteristics to desired levels. Over the last decade, much advances have been made in systems biology that allows system-wide characterization of cellular networks, both qualitatively and quantitatively, followed by whole-cell level engineering based on these findings. Furthermore, rapid advances in synthetic biology allow design and synthesis of fine controlled metabolic and gene regulatory circuits. The strategies and methods of systems biology and synthetic biology are rapidly integrated with metabolic engineering, thus resulting in "systems metabolic engineering".
In the paper published online in Nature Chemical Biology on May 17, Professor Sang Yup Lee and his colleagues at the Department of Chemical and Biomolecular Engineering, Korea Advanced Institute of Science and Technology (KAIST), Daejeon, Korea present new general strategies of systems metabolic engineering for developing microorganisms for the production of natural and non-natural chemicals from renewable biomass. They first classified the chemicals to be produced into four categories based on whether they have thus far been identified to exist in nature (natural vs. nonnatural) and whether they can be produced by inherent pathways of microorganisms (inherent, noninherent, or created): natural-inherent, natural-noninherent, non-natural-noninherent, and non-natural-created ones. General strategies for systems metabolic engineering of microorganisms for the production of these chemicals using various tools and methods based on omics, genome-scale metabolic modeling and simulation, evolutionary engineering, synthetic biology are suggested with relevant examples. For the production of non-natural chemicals, strategies for the construction of synthetic metabolic pathways are also suggested. Having collected diverse tools and methods for systems metabolic engineering, authors also suggest how to use them and their possible limitations.
Professor Sang Yup Lee said "It is expected that increasing number of chemicals and materials will be produced through biorefineries. We are now equipped with new strategies for developing microbial strains that can produce our desired products at very high efficiencies, thus allowing cost competitiveness to those produced by petrochemical refineries."
Editor of Nature Chemical Biology, Dr. Catherine Goodman, said "It is exciting to see how quickly science is progressing in this field – ideas that used to be science fiction are taking shape in research labs and biorefineries. The article by Professor Lee and his colleagues not only highlights the most advanced techniques and strategies available, but offers critical advice to progress the field as a whole."
The works of Professor Lee have been supported by the Advanced Biomass Center and Intelligent Synthetic Biology Center of Global Frontier Program from the Korean Ministry of Education, Science and Technology through National Research Foundation.
Contact: Dr. Sang Yup Lee, Distinguished Professor and Dean, KAIST, Daejeon, Korea (leesy@kaist.ac.kr, +82-42-350-3930)
2012.05.23 View 9644