research
- Multiple forms of a non-functional, unfolded protein follow different pathways and timelines to reach its folded, functional state, a study reveals. -

< Professor Hyotcherl Ihee (left), Professor Young Min Rhee (center), and Dr. Tae Wu Kim (right) >
KAIST researchers have used an X-ray method to track how proteins fold, which could improve computer simulations of this process, with implications for understanding diseases and improving drug discovery. Their findings were reported in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 30.
When proteins are translated from their DNA codes, they quickly transform from a non-functional, unfolded state into their folded, functional state. Problems in folding can lead to diseases like Alzheimer’s and Parkinson’s.
“Protein folding is one of the most important biological processes, as it forms the functioning 3D protein structure,” explained the physical chemist Hyotcherl Ihee of the Department of Chemistry at KAIST. Dr. Tae Wu Kim, the lead author of this research from Ihee’s group, added, “Understanding the mechanisms of protein folding is important, and could pave the way for disease study and drug development.”
Ihee’s team developed an approach using an X-ray scattering technique to uncover how the protein cytochrome c folds from its initial unfolded state. This protein is composed of a chain of 104 amino acids with an iron-containing heme molecule. It is often used for protein folding studies.
The researchers placed the protein in a solution and shined ultraviolet light on it. This process provides electrons to cytochrome c, reducing the iron within it from the ferric to the ferrous form, which initiates folding. As this was happening, the researchers beamed X-rays at very short intervals onto the sample. The X-rays scattered off all the atomic pairs in the sample and a detector continuously recorded the X-ray scattering patterns. The X-ray scattering patterns provided direct information regarding the 3D protein structure and the changes made in these patterns over time showed real-time motion of the protein during the folding process.
The team found cytochrome c proteins initially exist in a wide variety of unfolded states. Once the folding process is triggered, they stop by a group of intermediates within 31.6 microseconds, and then those intermediates follow different pathways with different folding times to reach an energetically stable folded state.
“We don’t know if this diversity in folding paths can be generalized to other proteins,” Ihee confessed. He continued, “However, we believe that our approach can be used to study other protein folding systems.”
Ihee hopes this approach can improve the accuracy of models that simulate protein interactions by including information on their unstructured states. These simulations are important as they can help identify barriers to proper folding and predict a protein’s folded state given its amino acid sequence. Ultimately, the models could help clarify how some diseases develop and how drugs interact with various protein structures.
Ihee’s group collaborated with Professor Young Min Rhee at the KAIST Department of Chemistry, and this work was supported by the National Research Foundation of Korea (NRF) and the Institute for Basic Science (IBS).
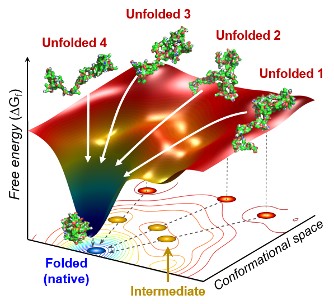
Figure. The scientists found that non-functional unfolded forms of the protein cytochrome c follow different pathways and timelines to reach a stable functional folded state.
Publications:
Kim, T. W., et al. (2020) ‘Protein folding from heterogeneous unfolded state revealed by time-resolved X-ray solution scattering’. PNAS. Volume 117. Issue 26. Page 14996-15005. Available online at https://doi.org/10.1073/pnas.1913442117
Profile: Hyotcherl Ihee, Ph.D.
Professor
hyotcherl.ihee@kaist.ac.kr
http://time.kaist.ac.kr/
Ihee Laboratory
Department of Chemistry
KAIST
https://www.kaist.ac.kr
Daejeon 34141, Korea
Profile: Young Min Rhee, Ph.D.
Professor
ymrhee@kaist.ac.kr
http://singlet.kaist.ac.kr
Rhee Research Group
Department of Chemistry
KAIST
https://www.kaist.ac.kr
Daejeon 34141, Korea
(END)
-
research KAIST Captures Protein Reaction in Just Six Milliseconds
Understanding biological phenomena, including protein-protein interactions and enzyme-substrate reactions occurring in microseconds to milliseconds, is essential for comprehending life processes and advancing drug development. KAIST researchers have developed a method for freezing and analyzing biochemical reaction changes within a span of just a few milliseconds, a critical step towards better understanding these complex biological reactions. < Photo. (From left) Professor Jin Young Kan
2025-03-24 -
research KAIST Uncovers the Principles of Gene Expression Regulation in Cancer and Cellular Functions
< (From left) Professor Seyun Kim, Professor Gwangrog Lee, Dr. Hyoungjoon Ahn, Dr. Jeongmin Yu, Professor Won-Ki Cho, and (below) PhD candidate Kwangmin Ryu of the Department of Biological Sciences> A research team at KAIST has identified the core gene expression networks regulated by key proteins that fundamentally drive phenomena such as cancer development, metastasis, tissue differentiation from stem cells, and neural activation processes. This discovery lays the foundation for dev
2025-01-24 -
research KAIST Develops CamBio - a New Biotemplating Method
- Professor Jae-Byum Chang and Professor Yeon Sik Jung’s joint research team of the Department of Materials Science and Engineering developed a highly tunable bio-templating method “CamBio” that makes use of intracellular protein structures - Substrate performance improvement of up to 230% demonstrated via surface-enhanced Raman spectroscopy (SERS) - Expected to have price competitiveness over bio-templating method as it expands the range of biological samples - Expected to
2025-01-10 -
research KAIST Researchers Suggest an Extraordinary Alternative to Petroleum-based PET - Bacteria!
< (From left) Dr. Cindy Pricilia, Ph.D. Candidate Cheon Woo Moon, Distinguished Professor Sang Yup Lee > Currently, the world is suffering from environmental problems caused by plastic waste. The KAIST research team has succeeded in producing a microbial-based plastic that is biodegradable and can replace existing PET bottles, making it a hot topic. Our university announced on the 7th of November that the research team of Distinguished Professor Sang Yup Lee of the Department of Ch
2024-11-08 -
research A KAIST Research Team Observes the Processes of Memory and Cognition in Real Time
The human brain contains approximately 86 billion neurons and 600 trillion synapses that exchange signals between the neurons to help us control the various functions of the brain including cognition, emotion, and memory. Interestingly, the number of synapses decrease with age or as a result of diseases like Alzheimer’s, and research on synapses thus attracts a lot of attention. However, limitations have existed in observing the dynamics of synapse structures in real time. On January 9,
2024-01-18