research
-Automated 3-D brain imaging data analysis technology offers more reliable and standardized analysis of the spatial organization of complex neural circuits.-
< PhD Candidate Jun Ho Song (left), Professor Se-Bum Paik (center), and PhD Candidate Woochul Choi (right) >,_Se-Bum_Paik_(center),_and_Woochul_Choi_(right).jpg)
KAIST researchers developed a new algorithm for brain imaging data analysis that enables the precise and quantitative mapping of complex neural circuits onto a standardized 3-D reference atlas.
Brain imaging data analysis is indispensable in the studies of neuroscience. However, analysis of obtained brain imaging data has been heavily dependent on manual processing, which cannot guarantee the accuracy, consistency, and reliability of the results.
Conventional brain imaging data analysis typically begins with finding a 2-D brain atlas image that is visually similar to the experimentally obtained brain image. Then, the region-of-interest (ROI) of the atlas image is matched manually with the obtained image, and the number of labeled neurons in the ROI is counted.
Such a visual matching process between experimentally obtained brain images and 2-D brain atlas images has been one of the major sources of error in brain imaging data analysis, as the process is highly subjective, sample-specific, and susceptible to human error. Manual analysis processes for brain images are also laborious, and thus studying the complete 3-D neuronal organization on a whole-brain scale is a formidable task.
To address these issues, a KAIST research team led by Professor Se-Bum Paik from the Department of Bio and Brain Engineering developed new brain imaging data analysis software named 'AMaSiNe (Automated 3-D Mapping of Single Neurons)', and introduced the algorithm in the May 26 issue of Cell Reports.
AMaSiNe automatically detects the positions of single neurons from multiple brain images, and accurately maps all the data onto a common standard 3-D reference space. The algorithm allows the direct comparison of brain data from different animals by automatically matching similar features from the images, and computing the image similarity score.
This feature-based quantitative image-to-image comparison technology improves the accuracy, consistency, and reliability of analysis results using only a small number of brain slice image samples, and helps standardize brain imaging data analyses.
Unlike other existing brain imaging data analysis methods, AMaSiNe can also automatically find the alignment conditions from misaligned and distorted brain images, and draw an accurate ROI, without any cumbersome manual validation process.
AMaSiNe has been further proved to produce consistent results with brain slice images stained utilizing various methods including DAPI, Nissl, and autofluorescence.
The two co-lead authors of this study, Jun Ho Song and Woochul Choi, exploited these benefits of AMaSiNe to investigate the topographic organization of neurons that project to the primary visual area (VISp) in various ROIs, such as the dorsal lateral geniculate nucleus (LGd), which could hardly be addressed without proper calibration and standardization of the brain slice image samples.
In collaboration with Professor Seung-Hee Lee's group of the Department of Biological Science, the researchers successfully observed the 3-D topographic neural projections to the VISp from LGd, and also demonstrated that these projections could not be observed when the slicing angle was not properly corrected by AMaSiNe. The results suggest that the precise correction of a slicing angle is essential for the investigation of complex and important brain structures.
AMaSiNe is widely applicable in the studies of various brain regions and other experimental conditions. For example, in the research team’s previous study jointly conducted with Professor Yang Dan’s group at UC Berkeley, the algorithm enabled the accurate analysis of the neuronal subsets in the substantia nigra and their projections to the whole brain. Their findings were published in Science on January 24.
AMaSiNe is of great interest to many neuroscientists in Korea and abroad, and is being actively used by a number of other research groups at KAIST, MIT, Harvard, Caltech, and UC San Diego.
Professor Paik said, “Our new algorithm allows the spatial organization of complex neural circuits to be found in a standardized 3-D reference atlas on a whole-brain scale. This will bring brain imaging data analysis to a new level.”
He continued, “More in-depth insights for understanding the function of brain circuits can be achieved by facilitating more reliable and standardized analysis of the spatial organization of neural circuits in various regions of the brain.”
This work was supported by KAIST and the National Research Foundation of Korea (NRF).
< Figure 1. Design of AMaSiNe to Overcome the Limits of Conventional Mouse Brain Imaging Data Analysis >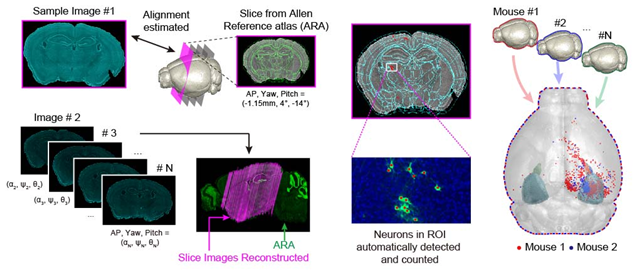
< Figure 2. Localization of Brain Slice Images onto the Standard Brain Atlas >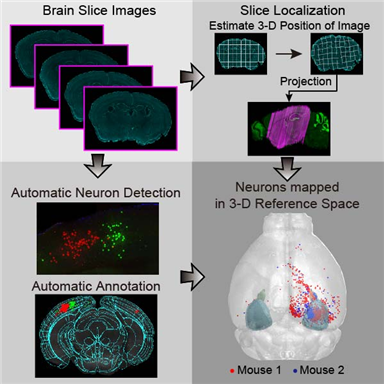
< Image. Standardized 3-D Mouse Brain >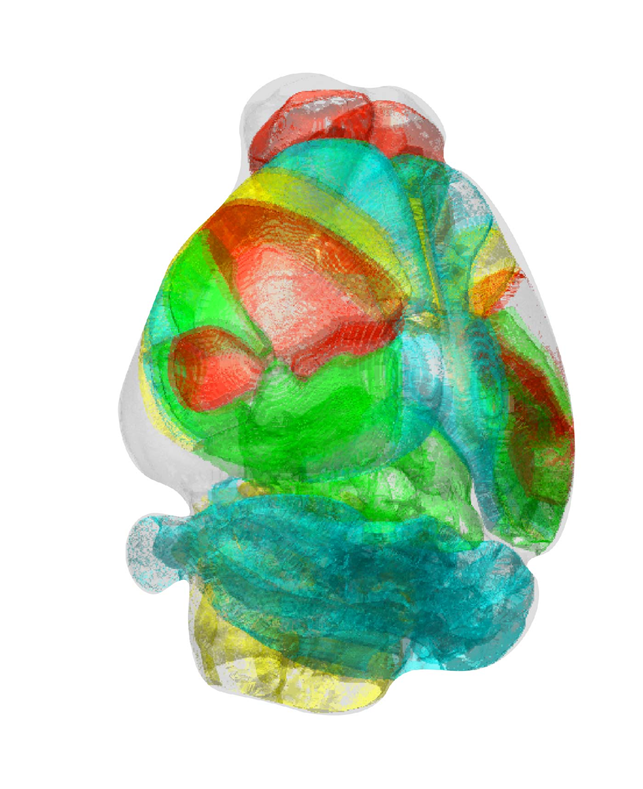
Figure and Image Credit: Professor Se-Bum Paik, KAIST
Figure and Image Usage Restrictions: News organizations may use or redistribute these figures and images, with proper attribution, as part of news coverage of this paper only.
Publication:
Song, J. H., et al. (2020). Precise Mapping of Single Neurons by Calibrated 3D Reconstruction of Brain Slices Reveals Topographic Projection in Mouse Visual Cortex. Cell Reports. Volume 31, 107682. Available online at https://doi.org/10.1016/j.celrep.2020.107682
Profile:
Se-Bum Paik
Assistant Professor
sbpaik@kaist.ac.kr
http://vs.kaist.ac.kr/
VSNN Laboratory
Department of Bio and Brain Engineering
Program of Brain and Cognitive Engineering
http://kaist.ac.kr
Korea Advanced Institute of Science and Technology (KAIST)
Daejeon, Republic of Korea
(END)
-
research KAIST Unveils New Possibilities for Treating Intractable Brain Tumors
< Photo 1. (From left) Professor Heung Kyu Lee, KAIST Department of Biological Sciences, and Dr. Keun Bon Ku > Immunotherapy, which enhances the immune system's T cell response to eliminate cancer cells, has emerged as a key approach in cancer treatment. However, in the case of glioblastoma, an aggressive and treatment-resistant brain tumor, numerous clinical trials have failed to confirm their efficacy. Korean researchers have recently analyzed the mechanisms that cause T cell exhaus
2024-11-15 -
research A KAIST research team identifies a cause of mental diseases induced by childhood abuse
Childhood neglect and/or abuse can induce extreme stress that significantly changes neural networks and functions during growth. This can lead to mental illnesses, including depression and schizophrenia, but the exact mechanism and means to control it were yet to be discovered. On August 1, a KAIST research team led by Professor Won-Suk Chung from the Department of Biological Sciences announced the identification of excessive synapse removal mediated by astrocytes as the cause of mental disea
2023-08-04 -
research The cause of disability in aged brain meningeal membranes identified
Due to the increase in average age, studies on changes in the brain following general aging process without serious brain diseases have also become an issue that requires in-depth studies. Regarding aging research, as aging progresses, ‘sugar’ accumulates in the body, and the accumulated sugar becomes a causative agent for various diseases such as aging-related inflammation and vascular disease. In the end, “surplus” sugar molecules attach to various proteins in the body
2023-03-15 -
people Professor Jae-Woong Jeong Receives Hyonwoo KAIST Academic Award
Professor Jae-Woong Jeong from the School of Electrical Engineering was selected for the Hyonwoo KAIST Academic Award, funded by the HyonWoo Cultural Foundation (Chairman Soo-il Kwak, honorary professor at Seoul National University Business School). The Hyonwoo KAIST Academic Award, presented for the first time in 2021, is an award newly founded by the donations of Chairman Soo-il Kwak of the HyonWoo Cultural Foundation, who aims to reward excellent KAIST scholars who have made outstanding a
2022-06-13 -
research Decoding Brain Signals to Control a Robotic Arm
Advanced brain-machine interface system successfully interprets arm movement directions from neural signals in the brain Researchers have developed a mind-reading system for decoding neural signals from the brain during arm movement. The method, described in the journal Applied Soft Computing, can be used by a person to control a robotic arm through a brain-machine interface (BMI). A BMI is a device that translates nerve signals into commands to control a machine, such as a computer or a rob
2022-03-18