research
A research group at KAIST presented a novel biosensor which can produce diverse, high-level microbial cell factories. The biosensor monitors the concentration of products and even intermediates when new strains are being developed. This strategy provides a new platform for manufacturing diverse natural products from renewable resources. The team succeeded in creating four natural products of high-level pharmaceutical importance with this strategy.
Malonyl-CoA is a major building block for many value-added chemicals including diverse natural products with pharmaceutical importance. However, due to the low availability of malonyl-CoA in bacteria, many malonyl-CoA-derived natural products have been produced by chemical synthesis or extraction from natural resources that are harmful to the environment and are unsustainable. For the sustainable biological production of malonyl-CoA-derived natural products, increasing the intracellular malonyl-CoA pool is necessary. To this end, the development of a robust and efficient malonyl-CoA biosensor was required to monitor the concentration of intracellular malonyl-CoA abundance as new strains are developed.
Metabolic engineering researchers at KAIST addressed this issue. This research reports the development of a simple and robust malonyl-CoA biosensor by repurposing a type III polyketide synthase (also known as RppA), which produces flaviolin, a colorimetric indicator of malonyl-CoA. Subsequently, the RppA biosensor was used for the rapid and efficient colorimetric screening of gene manipulation targets enabling enhanced malonyl-CoA abundance. The screened beneficial gene targets were employed for the high-level production of four representative natural products derived from malonyl-CoA. Compared with the previous strategies, which were expensive and time-consuming, the new biosensor could be easily applied to industrially relevant bacteria including Escherichia coli, Pseudomonas putida, and Corynebacterium glutamicum to enable a one-step process.
The study employs synthetic small regulatory RNA (sRNA) technology to rapidly and efficiently reduce endogenous target gene expression for improved malonyl-CoA production. The researchers constructed an E. coli genome-scale synthetic sRNA library targeting 1,858 genes covering all major metabolic genes in E. coli. This library was employed with the RppA biosensor to screen for gene targets which are believed to be beneficial for enhancing malonyl-CoA accumulation upon their expression knockdown.
From this colorimetric screening, 14 gene targets were selected, all of which were successful at significantly increasing the production of four natural products (6-methylsalicylic acid, aloesone, resveratrol, and naringenin). Although specific examples are demonstrated in E. coli as a host, the researchers showed that the biosensor is also functional in P. putida and C. glutamicum, industrially important representative gram-negative and gram-positive bacteria, respectively. The malonyl-CoA biosensor developed in this research will serve as an efficient platform for the rapid development of strains capable of producing natural products crucial for the pharmaceutical, chemical, cosmetics, and food industries.
An important aspect of this work is that the high-performance strains constructed in this research were developed rapidly and easily by utilizing the simple approach of colorimetric screening, without involving extensive metabolic engineering approaches. 6-Methylsalicylic acid (an antibiotic) could be produced to the highest titer reported for E. coli, and the microbial production of aloesone (a precursor of aloesin, an anti-inflammatory agent/whitening agent) was achieved for the first time.
“A sustainable process for producing diverse natural products using renewable resources is of great interest. This study represents the development of a robust and efficient malonyl-CoA biosensor generally applicable to a wide range of industrially important bacteria. The capability of this biosensor for screening a large library was demonstrated to show that the rapid and efficient construction of high-performance strains is feasible. This research will be useful for further accelerating the development process of strains capable of producing valuable chemicals to industrially relevant levels,” said Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering, who led the research.
This study entitled “Repurposing type III polyketide synthase as a malonyl-CoA biosensor for metabolic engineering in bacteria,” was published in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on October 02.
PhD students Dongsoo Yang and Won Jun Kim, MS student Shin Hee Ha, research staff Mun Hee Lee, Research Professor Seung Min Yoo, and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering and Dr. Jong Hyun Choi of the Applied Microbiology Research Center at the Korea Research Institute of Bioscience and Biotechnology (KRIBB) participated in this research.
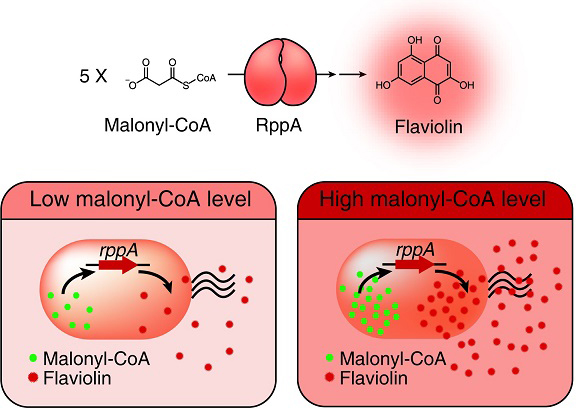
Figure: Type III polyketide synthase (RppA) as a malonyl-CoA biosensor. RppA converts five molecules of malonyl-CoA into one molecule of red-colored flaviolin. This schematic diagram shows the overall conceptualization of the malonyl-CoA biosensor by indicating that higher malonyl-CoA abundance leads to higher production and secretion of flaviolin, resulting in a deeper red color of the culture. This system was employed for the enhanced production of four representative natural products (6-methylsalicylic acid, aloesone, resveratrol, and naringenin) from engineered E. coli strains.
-
research KAIST Researchers Suggest an Extraordinary Alternative to Petroleum-based PET - Bacteria!
< (From left) Dr. Cindy Pricilia, Ph.D. Candidate Cheon Woo Moon, Distinguished Professor Sang Yup Lee > Currently, the world is suffering from environmental problems caused by plastic waste. The KAIST research team has succeeded in producing a microbial-based plastic that is biodegradable and can replace existing PET bottles, making it a hot topic. Our university announced on the 7th of November that the research team of Distinguished Professor Sang Yup Lee of the Department of Ch
2024-11-08 -
research X-ray Scattering Shines Light on Protein Folding
- Multiple forms of a non-functional, unfolded protein follow different pathways and timelines to reach its folded, functional state, a study reveals. - KAIST researchers have used an X-ray method to track how proteins fold, which could improve computer simulations of this process, with implications for understanding diseases and improving drug discovery. Their findings were reported in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 30. Whe
2020-07-09 -
research Recombinant E. Coli As a Biofactory for the Biosynthesis of Diverse Nanomaterials
(Distinguished Professor Lee and PhD candidate Choi) A metabolic research group at KAIST and Chung-Ang University in Korea has developed a recombinant E. coli strain that biosynthesizes 60 different nanomaterials covering 35 elements on the periodic table. Among the elements, the team could biosynthesize 33 novel nanomaterials for the first time, advancing the forward design of nanomaterials through the biosynthesis of various single and multi-elements. The study analy
2018-05-23 -
research Deep Learning Predicts Drug-Drug and Drug-Food Interactions
A Korean research team from KAIST developed a computational framework, DeepDDI, that accurately predicts and generates 86 types of drug-drug and drug-food interactions as outputs of human-readable sentences, which allows in-depth understanding of the drug-drug and drug-food interactions. Drug interactions, including drug-drug interactions (DDIs) and drug-food constituent interactions (DFIs), can trigger unexpected pharmacological effects, including adverse drug events (ADEs), wi
2018-04-18 -
research Professor Seyun Kim Identifies a Neuron Signal Controlling Molecule
A research team led by Professor Seyun Kim of the Department of Biological Sciences at KAIST has identified inositol pyrophosphates as the molecule that strongly controls neuron signaling via synaptotagmin. Professors Tae-Young Yoon of Yonsei University’s Y-IBS and Sung-Hyun Kim of Kyung Hee University’s Department of Biomedical Science also joined the team. The results were published in the Proceedings of the National Academy of Sciences of the United States of America (PN
2016-07-21