research
Researchers in Korea and the UK have successfully grown miniature models of critical lung structures called alveoli, and used them to study how the coronavirus that causes COVID-19 infects the lungs.
To date, there have been more than 40 million cases of COVID-19 and almost 1.13 million deaths worldwide. The main target tissues of SARS-CoV-2, the virus that causes COVID-19, especially in patients that develop pneumonia, appear to be alveoli – tiny air sacs in the lungs that take up the oxygen we breathe and exchange it with carbon dioxide to exhale.
To better understand how SARS-CoV-2 infects the lungs and causes disease, a team of Professor Young Seok Ju from the Graduate School of Medical Science and Engineering at KAIST in collaboration with the Wellcome-MRC Cambridge Stem Cell Institute at the University of Cambridge turned to organoids – ‘mini-organs’ grown in three dimensions to mimic the behaviour of tissue and organs.
The team used tissue donated to tissue banks at the Royal Papworth Hospital NHS Foundation Trust and Addenbrooke’s Hospital, Cambridge University NHS Foundations Trust, UK, and Seoul National University Hospital to extract a type of lung cell known as human lung alveolar type 2 cells. By reprogramming these cells back to their earlier ‘stem cell’ stage, they were able to grow self-organizing alveolar-like 3D structures that mimic the behaviour of key lung tissue.
“The research community now has a powerful new platform to study precisely how the virus infects the lungs, as well as explore possible treatments,” said Professor Ju, co-senior author of the research.
Dr. Joo-Hyeon Lee, another co-senior author at the Wellcome-MRC Cambridge Stem Cell Institute, said: “We still know surprisingly little about how SARS-CoV-2 infects the lungs and causes disease. Our approach has allowed us to grow 3D models of key lung tissue – in a sense, ‘mini-lungs’ – in the lab and study what happens when they become infected.”
The team infected the organoids with a strain of SARS-CoV-2 taken from a patient in Korea who was diagnosed with COVID-19 on January 26 after traveling to Wuhan, China. Using a combination of fluorescence imaging and single cell genetic analysis, they were able to study how the cells responded to the virus.
When the 3D models were exposed to SARS-CoV-2, the virus began to replicate rapidly, reaching full cellular infection just six hours after infection. Replication enables the virus to spread throughout the body, infecting other cells and tissue.
Around the same time, the cells began to produce interferons – proteins that act as warning signals to neighbouring cells, telling them to activate their antiviral defences. After 48 hours, the interferons triggered the innate immune response – its first line of defence – and the cells started fighting back against infection.
Sixty hours after infection, a subset of alveolar cells began to disintegrate, leading to cell death and damage to the lung tissue.
Although the researchers observed changes to the lung cells within three days of infection, clinical symptoms of COVID-19 rarely occur so quickly and can sometimes take more than ten days after exposure to appear. The team say there are several possible reasons for this. It may take several days from the virus first infiltrating the upper respiratory tract to it reaching the alveoli. It may also require a substantial proportion of alveolar cells to be infected or for further interactions with immune cells resulting in inflammation before a patient displays symptoms.
“Based on our model we can tackle many unanswered key questions, such as understanding genetic susceptibility to SARS-CoV-2, assessing relative infectivity of viral mutants, and revealing the damage processes of the virus in human alveolar cells,” said Professor Ju. “Most importantly, it provides the opportunity to develop and screen potential therapeutic agents against SARS-CoV-2 infection.”
“We hope to use our technique to grow these 3D models from cells of patients who are particularly vulnerable to infection, such as the elderly or people with diseased lungs, and find out what happens to their tissue,” added Dr. Lee.
The research was a collaboration involving scientists from KAIST, the University of Cambridge, Korea National Institute of Health, Institute for Basic Science (IBS), Seoul National University Hospital and Genome Insight in Korea.
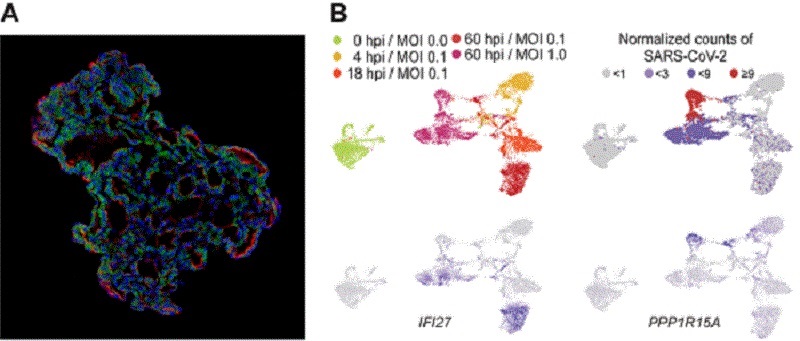
< (A)Representative image of three-dimensional human lung alveolar organoid showing alveolar stem cell marker, HTII-280 (red) and SARS-CoV-2 entry protein, ACE2 (green) (left panel). (B)Plot for single cell RNA sequencing. SARS-CoV-2 infected alveolar stem cells were heterogeneous: immune-proficient cells with moderate burden of SARS-CoV-2 and immune-deficient cells with high burden of SARS-CoV-2. hpi; hours post infection, MOI: multiplicity of infection, which means number of viral particles per a cell. >
- Profile
Professor Young Seok Ju
Laboratory of Cancer Genomics
https://julab.kaist.ac.kr
the Graduate School of Medical Science and Engineering
KAIST
- No Data