research
-
 KAIST's HUBO Ready for DARPA's Robotics Challenge Trials
When walking on muddy or bumpy roads, the two arms of DRC-HUBO become extra legs, enabling stable and agile movements.
The Humanoid Robot Research Center (HUBO Lab, http://hubolab.kaist.ac.kr) at the Korea Advanced Institute of Science and Technology (KAIST) and Rainbow Co., a spin-off venture company of the university, unveiled a new model of HUBO that will be entered in an international robotics competition scheduled later this year.
The competition is hosted and sponsored by the US Defense Advanced Research Projects Agency (DARPA), which is called the DARPA Robotics Challenge (DRC). Kicked off in October 2012, the DRC’s goal is to spur the development of advanced robots that can assist humans in mitigating and recovering from future natural and man-made disasters.
KAIST’s humanoid robot, HUBO, was originally created by Jun-Ho Oh, a distinguished professor of the Department of Mechanical Engineering, in 2004. Since then, the robot has gone through technological advancements, with the latest version of HUBO II released in 2012. So far, 12 HUBOs have been exported for further studies in robotics to universities, research institutes, and private companies in the US, China, and Singapore.
In tandem with Rainbow Co. (www.rainbow-robot.com), Professor Oh and his research team recently developed DRC-HUBO, which will compete as Team DRC-HUBO led by Drexel University at the DRC trials to be held in December 2013. Team DRC-HUBO is consisted of KAIST and nine US institutions.
DRC-HUBO is designed to perform difficult but essential activities required when responding to disaster scenes. The robot will have to fulfill eight tasks assigned by the DRC at the upcoming event such as driving a utility vehicle, walking across rough terrain, climbing a ladder, and using hand tools.
Unlike the previous models of HUBO, DRC-HUBO boasts several distinctive, enhanced features. Chief among them is the way the robot interacts with the external environment. Without complex sensors installed throughout the body, DRC-HUBO can control each joint of the arms and legs in compliance with the dynamics dictated by the external environment.
For example, when DRC-HUBO is faced with a rock falling from above while climbing up a ladder, the robot’s arms and legs naturally give in to the force of external changes. Accordingly, as the robot dodges the rock, its body and joints smoothly sway to absorb shock so that the fingers can keep a tight grip on the ladder, and the feet are planted firmly on the rail of the ladder, not losing balance.
In addition, DRC-HUBO can switch from bipedal to quadrupedal walking and vice versa. This provides the robot with greater stability to walk on uneven terrain or to climb up a hill. The robot’s arms and legs are elongated to better meet the challenges demanded by the DRC competition. DRC-HUBO’s two arms swing back and forth to form legs when necessary, thereby walking freely backwards and forwards.
The robot has gotten stronger grip as well. The right hand has four fingers (with one triggering finger that operates independently from the other three fingers), and the left hand has three fingers. All three fingers on both hands are actuated synchronously for gripping. The fingers are sophisticated enough to steer the wheel of a vehicle or grab a ladder to climb up, and strong enough to hold 15 lbs in one hand.
“With a full 34 degrees of freedom (DOF), DRC-HUBO stands 4.7 ft tall and weighs 120 lbs. All in all, the robot has been improved and extensively refurbished from the past models of HUBOs to compete at the DRC Trials. It has better vision and coordination. The legs and arms have become stronger,” said Professor Oh.
“Although the robot is still a prototype, it has important capabilities that can be utilized in advancing humanoid robots in general. One example is the way its arms can be used as extra legs to support the robot’s body, offering more flexibility in providing aid to humans.”
2013.07.25 View 13224
KAIST's HUBO Ready for DARPA's Robotics Challenge Trials
When walking on muddy or bumpy roads, the two arms of DRC-HUBO become extra legs, enabling stable and agile movements.
The Humanoid Robot Research Center (HUBO Lab, http://hubolab.kaist.ac.kr) at the Korea Advanced Institute of Science and Technology (KAIST) and Rainbow Co., a spin-off venture company of the university, unveiled a new model of HUBO that will be entered in an international robotics competition scheduled later this year.
The competition is hosted and sponsored by the US Defense Advanced Research Projects Agency (DARPA), which is called the DARPA Robotics Challenge (DRC). Kicked off in October 2012, the DRC’s goal is to spur the development of advanced robots that can assist humans in mitigating and recovering from future natural and man-made disasters.
KAIST’s humanoid robot, HUBO, was originally created by Jun-Ho Oh, a distinguished professor of the Department of Mechanical Engineering, in 2004. Since then, the robot has gone through technological advancements, with the latest version of HUBO II released in 2012. So far, 12 HUBOs have been exported for further studies in robotics to universities, research institutes, and private companies in the US, China, and Singapore.
In tandem with Rainbow Co. (www.rainbow-robot.com), Professor Oh and his research team recently developed DRC-HUBO, which will compete as Team DRC-HUBO led by Drexel University at the DRC trials to be held in December 2013. Team DRC-HUBO is consisted of KAIST and nine US institutions.
DRC-HUBO is designed to perform difficult but essential activities required when responding to disaster scenes. The robot will have to fulfill eight tasks assigned by the DRC at the upcoming event such as driving a utility vehicle, walking across rough terrain, climbing a ladder, and using hand tools.
Unlike the previous models of HUBO, DRC-HUBO boasts several distinctive, enhanced features. Chief among them is the way the robot interacts with the external environment. Without complex sensors installed throughout the body, DRC-HUBO can control each joint of the arms and legs in compliance with the dynamics dictated by the external environment.
For example, when DRC-HUBO is faced with a rock falling from above while climbing up a ladder, the robot’s arms and legs naturally give in to the force of external changes. Accordingly, as the robot dodges the rock, its body and joints smoothly sway to absorb shock so that the fingers can keep a tight grip on the ladder, and the feet are planted firmly on the rail of the ladder, not losing balance.
In addition, DRC-HUBO can switch from bipedal to quadrupedal walking and vice versa. This provides the robot with greater stability to walk on uneven terrain or to climb up a hill. The robot’s arms and legs are elongated to better meet the challenges demanded by the DRC competition. DRC-HUBO’s two arms swing back and forth to form legs when necessary, thereby walking freely backwards and forwards.
The robot has gotten stronger grip as well. The right hand has four fingers (with one triggering finger that operates independently from the other three fingers), and the left hand has three fingers. All three fingers on both hands are actuated synchronously for gripping. The fingers are sophisticated enough to steer the wheel of a vehicle or grab a ladder to climb up, and strong enough to hold 15 lbs in one hand.
“With a full 34 degrees of freedom (DOF), DRC-HUBO stands 4.7 ft tall and weighs 120 lbs. All in all, the robot has been improved and extensively refurbished from the past models of HUBOs to compete at the DRC Trials. It has better vision and coordination. The legs and arms have become stronger,” said Professor Oh.
“Although the robot is still a prototype, it has important capabilities that can be utilized in advancing humanoid robots in general. One example is the way its arms can be used as extra legs to support the robot’s body, offering more flexibility in providing aid to humans.”
2013.07.25 View 13224 -
 A magnetic pen for smartphones adds another level of conveniences
Utilizing existing features on smartphones, the MagPen provides users with a compatible and simple input tool regardless of the type of phones they are using.
A doctoral candidate at the Korea Advanced Institute of Science and Technology (KAIST) developed a magnetically driven pen interface that works both on and around mobile devices. This interface, called the MagPen, can be used for any type of smartphones and tablet computers so long as they have magnetometers embedded in.
Advised by Professor Kwang-yun Wohn of the Graduate School of Culture Technology (GSCT) at KAIST, Sungjae Hwang, a Ph.D. student, created the MagPen in collaboration with Myung-Wook Ahn, a master"s student at the GSCT of KAIST, and Andrea Bianchi, a professor at Sungkyunkwan University.
Almost all mobile devices today provide location-based services, and magnetometers are incorporated in the integrated circuits of smartphones or tablet PCs, functioning as compasses. Taking advantage of built-in magnetometers, Hwang"s team came up with a technology that enabled an input tool for mobile devices such as a capacitive stylus pen to interact more sensitively and effectively with the devices" touch screen. Text and command entered by a stylus pen are expressed better on the screen of mobile devices than those done by human fingers.
The MagPen utilizes magnetometers equipped with smartphones, thus there is no need to build an additional sensing panel for a touchscreen as well as circuits, communication modules, or batteries for the pen. With an application installed on smartphones, it senses and analyzes the magnetic field produced by a permanent magnet embedded in a standard capacitive stylus pen.
Sungjae Hwang said, "Our technology is eco-friendly and very affordable because we are able to improve the expressiveness of the stylus pen without requiring additional hardware beyond those already installed on the current mobile devices. The technology allows smartphone users to enjoy added convenience while no wastes generated."
The MagPen detects the direction at which a stylus pen is pointing; selects colors by dragging the pen across smartphone bezel; identifies pens with different magnetic properties; recognizes pen-spinning gestures; and estimates the finger pressure applied to the pen.
Notably, with its spinning motion, the MagPen expands the scope of input gestures recognized by a stylus pen beyond its existing vocabularies of gestures and techniques such as titling, hovering, and varying pressures. The tip of the pen switches from a pointer to an eraser and vice versa when spinning. Or, it can choose the thickness of the lines drawn on a screen by spinning.
"It"s quite remarkable to see that the MagPen can understand spinning motion. It"s like the pen changes its living environment from two dimensions to three dimensions. This is the most creative characteristic of our technology," added Sungjae Hwang.
Hwang"s initial research result was first presented at the International Conference on Intelligent User Interfaces organized by the Association for Computing Machinery and held on March 19-22 in Santa Monica, the US.
In the next month of August, the research team will present a paper on the MagPen technology, entitled "MagPen: Magnetically Driven Pen Interaction On and Around Conventional Smartphones" and receive an Honorable Mention Award at the 15th International Conference on Human-Computer Interaction with Mobile Devices and Services (MobileHCI 2013) to be held in Germany.
In addition to the MagPen, Hwang and his team are conducting other projects to develop different types of magnetic gadgets (collectively called "MagGetz") that include the Magnetic Marionette, a magnetic cover for a smartphone, which offers augmented interactions with the phone, as well as magnetic widgets such as buttons and toggle interface.
Hwang has filed ten patents for the MagGetz technology.
Youtube Links: http://www.youtube.com/watch?v=NkPo2las7wc, http://www.youtube.com/watch?v=J9GtgyzoZmM
2013.07.25 View 9784
A magnetic pen for smartphones adds another level of conveniences
Utilizing existing features on smartphones, the MagPen provides users with a compatible and simple input tool regardless of the type of phones they are using.
A doctoral candidate at the Korea Advanced Institute of Science and Technology (KAIST) developed a magnetically driven pen interface that works both on and around mobile devices. This interface, called the MagPen, can be used for any type of smartphones and tablet computers so long as they have magnetometers embedded in.
Advised by Professor Kwang-yun Wohn of the Graduate School of Culture Technology (GSCT) at KAIST, Sungjae Hwang, a Ph.D. student, created the MagPen in collaboration with Myung-Wook Ahn, a master"s student at the GSCT of KAIST, and Andrea Bianchi, a professor at Sungkyunkwan University.
Almost all mobile devices today provide location-based services, and magnetometers are incorporated in the integrated circuits of smartphones or tablet PCs, functioning as compasses. Taking advantage of built-in magnetometers, Hwang"s team came up with a technology that enabled an input tool for mobile devices such as a capacitive stylus pen to interact more sensitively and effectively with the devices" touch screen. Text and command entered by a stylus pen are expressed better on the screen of mobile devices than those done by human fingers.
The MagPen utilizes magnetometers equipped with smartphones, thus there is no need to build an additional sensing panel for a touchscreen as well as circuits, communication modules, or batteries for the pen. With an application installed on smartphones, it senses and analyzes the magnetic field produced by a permanent magnet embedded in a standard capacitive stylus pen.
Sungjae Hwang said, "Our technology is eco-friendly and very affordable because we are able to improve the expressiveness of the stylus pen without requiring additional hardware beyond those already installed on the current mobile devices. The technology allows smartphone users to enjoy added convenience while no wastes generated."
The MagPen detects the direction at which a stylus pen is pointing; selects colors by dragging the pen across smartphone bezel; identifies pens with different magnetic properties; recognizes pen-spinning gestures; and estimates the finger pressure applied to the pen.
Notably, with its spinning motion, the MagPen expands the scope of input gestures recognized by a stylus pen beyond its existing vocabularies of gestures and techniques such as titling, hovering, and varying pressures. The tip of the pen switches from a pointer to an eraser and vice versa when spinning. Or, it can choose the thickness of the lines drawn on a screen by spinning.
"It"s quite remarkable to see that the MagPen can understand spinning motion. It"s like the pen changes its living environment from two dimensions to three dimensions. This is the most creative characteristic of our technology," added Sungjae Hwang.
Hwang"s initial research result was first presented at the International Conference on Intelligent User Interfaces organized by the Association for Computing Machinery and held on March 19-22 in Santa Monica, the US.
In the next month of August, the research team will present a paper on the MagPen technology, entitled "MagPen: Magnetically Driven Pen Interaction On and Around Conventional Smartphones" and receive an Honorable Mention Award at the 15th International Conference on Human-Computer Interaction with Mobile Devices and Services (MobileHCI 2013) to be held in Germany.
In addition to the MagPen, Hwang and his team are conducting other projects to develop different types of magnetic gadgets (collectively called "MagGetz") that include the Magnetic Marionette, a magnetic cover for a smartphone, which offers augmented interactions with the phone, as well as magnetic widgets such as buttons and toggle interface.
Hwang has filed ten patents for the MagGetz technology.
Youtube Links: http://www.youtube.com/watch?v=NkPo2las7wc, http://www.youtube.com/watch?v=J9GtgyzoZmM
2013.07.25 View 9784 -
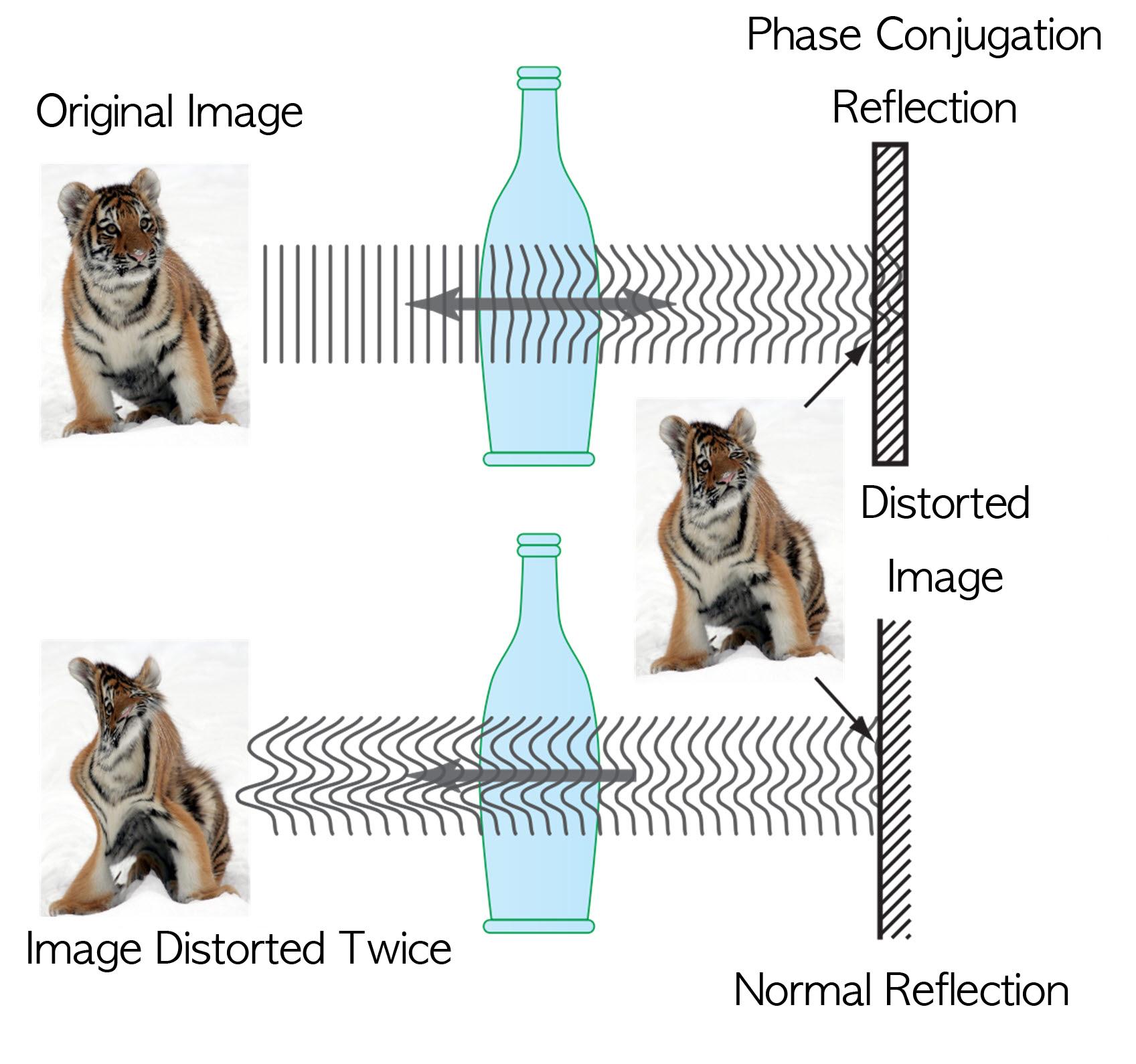 Technology Developed to Control Light Scattering Using Holography
Published on May 29th Nature Scientific Reports online
Recently, a popular article demonstrated that an opaque glass becomes transparent as transparent tape is applied to the glass. The scientific principle is that light is less scattered as the rough surface of the opaque glass is filled by transparent tape, thereby making things behind the opaque glass look clearer.
Professor Yong-Keun Park from KAIST’s Department of Physics, in a joint research with MIT Spectroscopy Lab, has developed a technology to easily control light scattering using holography. Their results are published on Nature’s Scientific Reports May 29th online edition.
This technology allows us to see things behind visual obstructions such as cloud and smoke, or even human skin that is highly scattering, optically thick materials. The research team applied the holography technology that records both the direction and intensity of light, and controlled light scattering of obstacles lied between an observer and a target image. The team was able to retrieve the original image by recording the information of scattered light and reflecting the light precisely to the other side.This phenomenon is known as “phase conjugation” in physics. Professor Park’s team applied phase conjugation and digital holography to observe two-dimensional image behind a highly scattering wall. “This technology will be utilized in many fields of physics, optics, nanotechnology, medical science, and even military science,” said Professor Park. “This is different from what is commonly known as penetrating camera or invisible clothes.” He nevertheless drew the line at over-interpreting the technology, “Currently, the significance is on the development of the technology itself that allows us to accurately control the scattering of light."
Figure I. Observed Images
Figure II. Light Scattering Control
2013.07.19 View 7699
Technology Developed to Control Light Scattering Using Holography
Published on May 29th Nature Scientific Reports online
Recently, a popular article demonstrated that an opaque glass becomes transparent as transparent tape is applied to the glass. The scientific principle is that light is less scattered as the rough surface of the opaque glass is filled by transparent tape, thereby making things behind the opaque glass look clearer.
Professor Yong-Keun Park from KAIST’s Department of Physics, in a joint research with MIT Spectroscopy Lab, has developed a technology to easily control light scattering using holography. Their results are published on Nature’s Scientific Reports May 29th online edition.
This technology allows us to see things behind visual obstructions such as cloud and smoke, or even human skin that is highly scattering, optically thick materials. The research team applied the holography technology that records both the direction and intensity of light, and controlled light scattering of obstacles lied between an observer and a target image. The team was able to retrieve the original image by recording the information of scattered light and reflecting the light precisely to the other side.This phenomenon is known as “phase conjugation” in physics. Professor Park’s team applied phase conjugation and digital holography to observe two-dimensional image behind a highly scattering wall. “This technology will be utilized in many fields of physics, optics, nanotechnology, medical science, and even military science,” said Professor Park. “This is different from what is commonly known as penetrating camera or invisible clothes.” He nevertheless drew the line at over-interpreting the technology, “Currently, the significance is on the development of the technology itself that allows us to accurately control the scattering of light."
Figure I. Observed Images
Figure II. Light Scattering Control
2013.07.19 View 7699 -
 Nanofiber sensor detects diabetes or lung cancer faster and easier
Metal-oxide nanofiber based chemiresistive gas sensors offer greater usability for portable real-time breath tests that can be available on smart phones or tablet PCs in the near future.
Daejeon, Republic of Korea, June 11, 2013 -- Today"s technological innovation enables smartphone users to diagnose serious diseases such as diabetes or lung cancer quickly and effectively by simply breathing into a small gadget, a nanofiber breathing sensor, mounted on the phones.
Il-Doo Kim, Associate Professor of Materials Science and Engineering Department at the Korea Advanced Institute of Science and Technology (KAIST), and his research team have recently published a cover paper entitled "Thin-Wall Assembled SnO2 Fibers Functionalized by Catalytic Pt Nanoparticles and their Superior Exhaled Breath-Sensing Properties for the Diagnosis of Diabetes," in an academic journal, Advanced Functional Materials (May 20th issue), on the development of a highly sensitive exhaled breath sensor by using hierarchical SnO2 fibers that are assembled from wrinkled thin SnO2 nanotubes.
In the paper, the research team presented a morphological evolution of SnO2 fibers, called micro phase-separations, which takes place between polymers and other dissolved solutes when varying the flow rate of an electrospinning solution feed and applying a subsequent heat treatment afterward.
The morphological change results in nanofibers that are shaped like an open cylinder inside which thin-film SnO2 nanotubes are layered and then rolled up. A number of elongated pores ranging from 10 nanometers (nm) to 500 nm in length along the fiber direction were formed on the surface of the SnO2 fibers, allowing exhaled gas molecules to easily permeate the fibers. The inner and outer wall of SnO2 tubes is evenly coated with catalytic platinum (Pt) nanoparticles. According to the research team, highly porous SnO2 fibers, synthesized by eletrospinning at a high flow rate, showed five-fold higher acetone responses than that of the dense SnO2 nanofibers created under a low flow rate. The catalytic Pt coating shortened the fibers" gas response time dramatically as well.
The breath analysis for diabetes is largely based on an acetone breath test because acetone is one of the specific volatile organic compounds (VOC) produced in the human body to signal the onset of particular diseases. In other words, they are biomarkers to predict certain diseases such as acetone for diabetes, toluene for lung cancer, and ammonia for kidney malfunction. Breath analysis for medical evaluation has attracted much attention because it is less intrusive than conventional medical examination, as well as fast and convenient, and environmentally friendly, leaving almost no biohazard wastes.
Various gas-sensing techniques have been adopted to analyze VOCs including gas chromatography-mass spectroscopy (GC-MS), but these techniques are difficult to incorporate into portable real-time gas sensors because the testing equipment is bulky and expensive, and their operation is more complex. Metal-oxide based chemiresistive gas sensors, however, offer greater usability for portable real-time breath sensors.
Il-Doo Kim said, "Catalyst-loaded metal oxide nanofibers synthesized by electrospinning have a great potential for future exhaled breath sensor applications. From our research, we obtained the results that Pt-coated SnO2 fibers are able to identify promptly and accurately acetone or toluene even at very low concentration less than 100 parts per billion (ppb)."
The exhaled acetone level of diabetes patients exceeds 1.8 parts per million (ppm), which is two to six-fold higher than that (0.3-0.9 ppm) of healthy people. Therefore, a highly sensitive detection that responds to acetone below 1 ppm, in the presence of other exhaled gases as well as under the humid environment of human breath, is important for an accurate diagnosis of diabetes. In addition, Professor Kim said, "a trace concentration of toluene (30 ppb) in exhaled breath is regarded to be a distinctive early symptom of lung cancer, which we were able to detect with our prototype breath tester."
The research team has now been developing an array of breathing sensors using various catalysts and a number of semiconducting metal oxide fibers, which will offer patients a real-time easy diagnosis of diseases.
###
Youtube Link: http://www.youtube.com/watch?v=t_Hr11dRryg
For further inquires:
Il-Doo Kim, Professor of Materials Science and Engineering, KAIST
Advanced Nanomaterials and Energy Laboratory
Tel: +82-42-350-3329
Email: idkim@kaist.ac.kr
Clockwise from left to right: left upper shows a magnified SEM image of a broken thin-wall assembled SnO2 fiber. Left below is an array of breath sensors (Inset is an actual size of a breath sensor). The right is the cover of Advanced Functional Materials (May 20th issue) in which a research paper on the development of a highly sensitive exhaled breath sensor by using SnO2 fibers is published.
This is the microstructural evolution of SnO2 nanofibers as a function of flow rate during electrospinning.
2013.06.20 View 13283
Nanofiber sensor detects diabetes or lung cancer faster and easier
Metal-oxide nanofiber based chemiresistive gas sensors offer greater usability for portable real-time breath tests that can be available on smart phones or tablet PCs in the near future.
Daejeon, Republic of Korea, June 11, 2013 -- Today"s technological innovation enables smartphone users to diagnose serious diseases such as diabetes or lung cancer quickly and effectively by simply breathing into a small gadget, a nanofiber breathing sensor, mounted on the phones.
Il-Doo Kim, Associate Professor of Materials Science and Engineering Department at the Korea Advanced Institute of Science and Technology (KAIST), and his research team have recently published a cover paper entitled "Thin-Wall Assembled SnO2 Fibers Functionalized by Catalytic Pt Nanoparticles and their Superior Exhaled Breath-Sensing Properties for the Diagnosis of Diabetes," in an academic journal, Advanced Functional Materials (May 20th issue), on the development of a highly sensitive exhaled breath sensor by using hierarchical SnO2 fibers that are assembled from wrinkled thin SnO2 nanotubes.
In the paper, the research team presented a morphological evolution of SnO2 fibers, called micro phase-separations, which takes place between polymers and other dissolved solutes when varying the flow rate of an electrospinning solution feed and applying a subsequent heat treatment afterward.
The morphological change results in nanofibers that are shaped like an open cylinder inside which thin-film SnO2 nanotubes are layered and then rolled up. A number of elongated pores ranging from 10 nanometers (nm) to 500 nm in length along the fiber direction were formed on the surface of the SnO2 fibers, allowing exhaled gas molecules to easily permeate the fibers. The inner and outer wall of SnO2 tubes is evenly coated with catalytic platinum (Pt) nanoparticles. According to the research team, highly porous SnO2 fibers, synthesized by eletrospinning at a high flow rate, showed five-fold higher acetone responses than that of the dense SnO2 nanofibers created under a low flow rate. The catalytic Pt coating shortened the fibers" gas response time dramatically as well.
The breath analysis for diabetes is largely based on an acetone breath test because acetone is one of the specific volatile organic compounds (VOC) produced in the human body to signal the onset of particular diseases. In other words, they are biomarkers to predict certain diseases such as acetone for diabetes, toluene for lung cancer, and ammonia for kidney malfunction. Breath analysis for medical evaluation has attracted much attention because it is less intrusive than conventional medical examination, as well as fast and convenient, and environmentally friendly, leaving almost no biohazard wastes.
Various gas-sensing techniques have been adopted to analyze VOCs including gas chromatography-mass spectroscopy (GC-MS), but these techniques are difficult to incorporate into portable real-time gas sensors because the testing equipment is bulky and expensive, and their operation is more complex. Metal-oxide based chemiresistive gas sensors, however, offer greater usability for portable real-time breath sensors.
Il-Doo Kim said, "Catalyst-loaded metal oxide nanofibers synthesized by electrospinning have a great potential for future exhaled breath sensor applications. From our research, we obtained the results that Pt-coated SnO2 fibers are able to identify promptly and accurately acetone or toluene even at very low concentration less than 100 parts per billion (ppb)."
The exhaled acetone level of diabetes patients exceeds 1.8 parts per million (ppm), which is two to six-fold higher than that (0.3-0.9 ppm) of healthy people. Therefore, a highly sensitive detection that responds to acetone below 1 ppm, in the presence of other exhaled gases as well as under the humid environment of human breath, is important for an accurate diagnosis of diabetes. In addition, Professor Kim said, "a trace concentration of toluene (30 ppb) in exhaled breath is regarded to be a distinctive early symptom of lung cancer, which we were able to detect with our prototype breath tester."
The research team has now been developing an array of breathing sensors using various catalysts and a number of semiconducting metal oxide fibers, which will offer patients a real-time easy diagnosis of diseases.
###
Youtube Link: http://www.youtube.com/watch?v=t_Hr11dRryg
For further inquires:
Il-Doo Kim, Professor of Materials Science and Engineering, KAIST
Advanced Nanomaterials and Energy Laboratory
Tel: +82-42-350-3329
Email: idkim@kaist.ac.kr
Clockwise from left to right: left upper shows a magnified SEM image of a broken thin-wall assembled SnO2 fiber. Left below is an array of breath sensors (Inset is an actual size of a breath sensor). The right is the cover of Advanced Functional Materials (May 20th issue) in which a research paper on the development of a highly sensitive exhaled breath sensor by using SnO2 fibers is published.
This is the microstructural evolution of SnO2 nanofibers as a function of flow rate during electrospinning.
2013.06.20 View 13283 -
 Technology for Non-Breaking Smartphone Display Developed
High-strength plastic display has been developed by applying a glass-fiber fabric.
“Will bring about innovation to the field by replacing glass substrates”
It is now possible to manufacture non-breaking smartphone display. Heavy glass substrates of large-screen televisions will be replaced with light plastic films.
Professor Choon Sup Yoon from KAIST’s Department of Physics and KAIST Institute for Information Technology Convergence has developed the technology for high-strength plastic substrates to replace glass displays.
The plastic substrate created by Professor Yoon and his research team have greatly enhanced needed properties of heat resistance, transparency, flexibility, inner chemical capability, and tensile strength. Although the material retains flexibility as a native advantage of plastic film, its tensile strength is three times greater than that of normal glass, which is a degree similar to tempered glass. In addition, Professor Yoon’s substrate is as colorless and transparent as glass and resists heat up to 450℃, while its thermal expansivity is only 10% to 20% of existing plastics.
Glass substrates are currently used in practically every display such as mobile phone screens, televisions, and computer monitors for having smooth surface and satisfying basic conditions for display substrates. However, as glass substrates are heavy and easily broken, researchers studied colorless and transparent plastic polyimide films to replace glass substrates for their excellent thermal and chemical stability.
Nonetheless, colorless and transparent polyimide films do not have sufficient heat resistance and mechanical solidity. To resolve this problem, polyimide films are impregnated with glass-fiber fabrics, but it was far from commercialization as the impregnation exacerbates the roughness of surface and light transmittance. The roughness of the surface increases as the solvent evaporates in the impregnation process, resulting in surface roughness of around 0.4μm. The downturn in light transmittance is due to light scattering effect by the discording refractive index of polyimide film and glass-fiber fabric.
Professor Yoon’s research team resolved these issues by tuning the refractive indices of transparent polyimide film and glass-fiber fabric up to four decimal places, and by developing the technology of flattening the film’s surface roughness to a few nanometers. As a result, the research team achieved heat expansivity of 11ppm/℃, surface roughness of 0.9nm, tensile strength of 250MPa, bending curvature radius of 2mm, and light transmittance at 90% with a 110μm-thick glass-fiber fabric impregnated transparent polyimide film substrate.
“The developed substrate can not only replace the traditional glass substrate but also be applied as flexible display substrate,” said Professor Yoon in prospect, “it will bring about technological innovation in display industry as it can fundamentally resolve the issue of shattering mobile phone displays, reduce the weight and thickness of large-area televisions, and apply Roll to Roll process in display manufacture.”
Supported by the Ministry of Knowledge Economy for five years, the technology has applied for 3 patents and is in discussion for technology transfer with related business.
Figure 1. The according (left) and discording (right) refractive indices of glass-fiber fabric and polyimide film. The characters on the left are sharp and clear, but the characters on the right appear foggy.
Figure 2. Picture of the developed glass-fiber fabric
2013.06.09 View 8417
Technology for Non-Breaking Smartphone Display Developed
High-strength plastic display has been developed by applying a glass-fiber fabric.
“Will bring about innovation to the field by replacing glass substrates”
It is now possible to manufacture non-breaking smartphone display. Heavy glass substrates of large-screen televisions will be replaced with light plastic films.
Professor Choon Sup Yoon from KAIST’s Department of Physics and KAIST Institute for Information Technology Convergence has developed the technology for high-strength plastic substrates to replace glass displays.
The plastic substrate created by Professor Yoon and his research team have greatly enhanced needed properties of heat resistance, transparency, flexibility, inner chemical capability, and tensile strength. Although the material retains flexibility as a native advantage of plastic film, its tensile strength is three times greater than that of normal glass, which is a degree similar to tempered glass. In addition, Professor Yoon’s substrate is as colorless and transparent as glass and resists heat up to 450℃, while its thermal expansivity is only 10% to 20% of existing plastics.
Glass substrates are currently used in practically every display such as mobile phone screens, televisions, and computer monitors for having smooth surface and satisfying basic conditions for display substrates. However, as glass substrates are heavy and easily broken, researchers studied colorless and transparent plastic polyimide films to replace glass substrates for their excellent thermal and chemical stability.
Nonetheless, colorless and transparent polyimide films do not have sufficient heat resistance and mechanical solidity. To resolve this problem, polyimide films are impregnated with glass-fiber fabrics, but it was far from commercialization as the impregnation exacerbates the roughness of surface and light transmittance. The roughness of the surface increases as the solvent evaporates in the impregnation process, resulting in surface roughness of around 0.4μm. The downturn in light transmittance is due to light scattering effect by the discording refractive index of polyimide film and glass-fiber fabric.
Professor Yoon’s research team resolved these issues by tuning the refractive indices of transparent polyimide film and glass-fiber fabric up to four decimal places, and by developing the technology of flattening the film’s surface roughness to a few nanometers. As a result, the research team achieved heat expansivity of 11ppm/℃, surface roughness of 0.9nm, tensile strength of 250MPa, bending curvature radius of 2mm, and light transmittance at 90% with a 110μm-thick glass-fiber fabric impregnated transparent polyimide film substrate.
“The developed substrate can not only replace the traditional glass substrate but also be applied as flexible display substrate,” said Professor Yoon in prospect, “it will bring about technological innovation in display industry as it can fundamentally resolve the issue of shattering mobile phone displays, reduce the weight and thickness of large-area televisions, and apply Roll to Roll process in display manufacture.”
Supported by the Ministry of Knowledge Economy for five years, the technology has applied for 3 patents and is in discussion for technology transfer with related business.
Figure 1. The according (left) and discording (right) refractive indices of glass-fiber fabric and polyimide film. The characters on the left are sharp and clear, but the characters on the right appear foggy.
Figure 2. Picture of the developed glass-fiber fabric
2013.06.09 View 8417 -
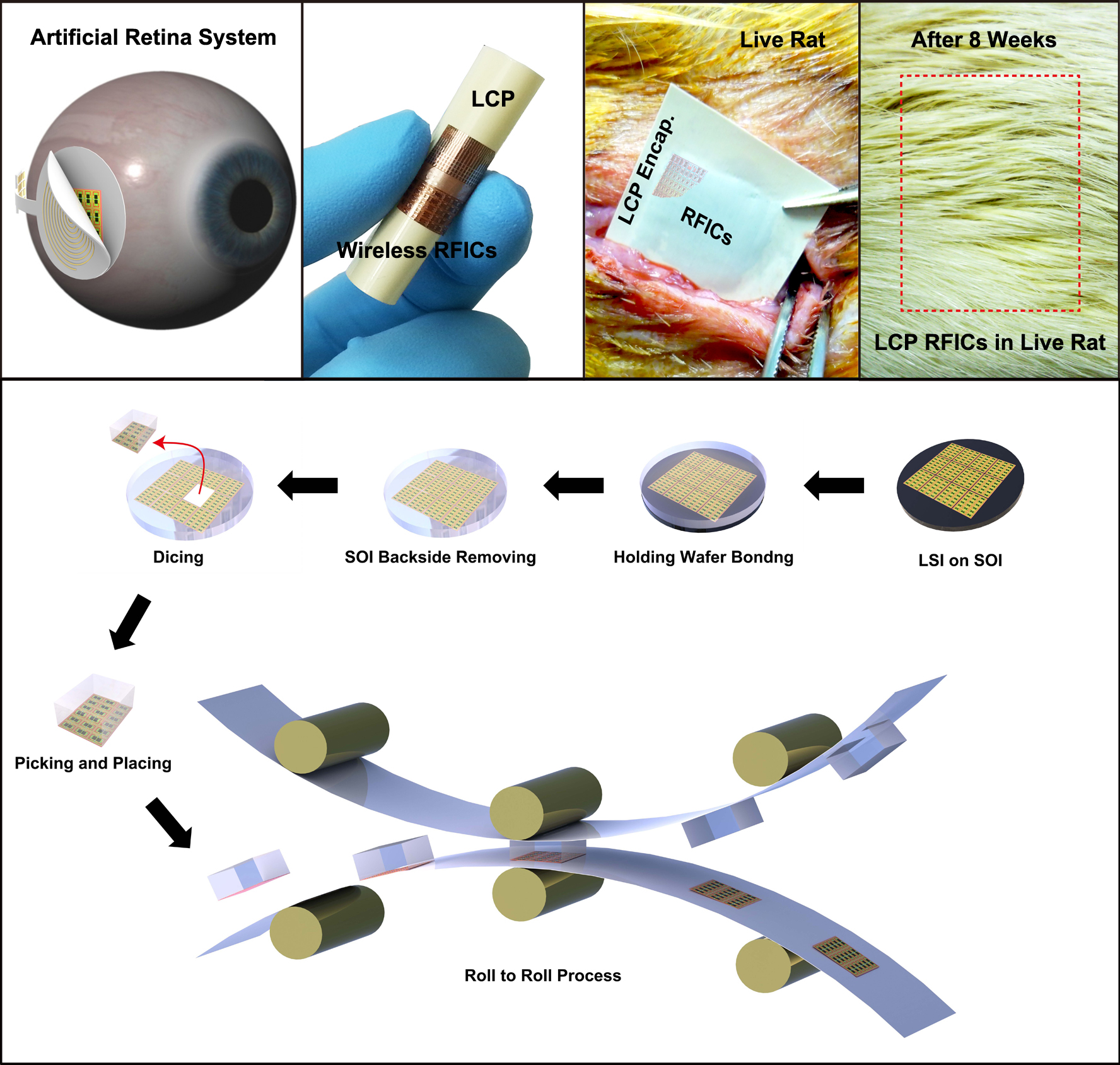 A KAIST research team developed in vivo flexible large scale integrated circuits
Daejeon, Republic of Korea, May 6th, 2013–-A team led by Professor Keon Jae Lee from the Department of Materials Science and Engineering at KAIST has developed in vivo silicon-based flexible large scale integrated circuits (LSI) for bio-medical wireless communication.
Silicon-based semiconductors have played significant roles in signal processing, nerve stimulation, memory storage, and wireless communication in implantable electronics. However, the rigid and bulky LSI chips have limited uses in in vivo devices due to incongruent contact with the curvilinear surfaces of human organs. Especially, artificial retinas recently approved by the Food and Drug Administration (refer to the press release of FDA"s artificial retina approval) require extremely flexible and slim LSI to incorporate it within the cramped area of the human eye.
Although several research teams have fabricated flexible integrated circuits (ICs, tens of interconnected transistors) on plastics, their inaccurate nano-scale alignment on plastics has restricted the demonstration of flexible nano-transistors and their large scale interconnection for in vivo LSI applications such as main process unit (MPU), high density memory and wireless communication. Professor Lee"s team previously demonstrated fully functional flexible memory using ultrathin silicon membranes (Nano Letters, Flexible Memristive Memory Array on Plastic Substrates), however, its integration level and transistor size (over micron scale) have limited functional applications for flexible consumer electronics.
Professor Keon Jae Lee"s team fabricated radio frequency integrated circuits (RFICs) interconnected with thousand nano-transistors on silicon wafer by state-of-the-art CMOS process, and then they removed the entire bottom substrate except top 100 nm active circuit layer by wet chemical etching. The flexible RF switches for wireless communication were monolithically encapsulated with biocompatible liquid crystal polymers (LCPs) for in vivo bio-medical applications. Finally, they implanted the LCP encapsulated RFICs into live rats to demonstrate the stable operation of flexible devices under in vivo circumstances.
Professor Lee said, "This work could provide an approach to flexible LSI for an ideal artificial retina system and other bio-medical devices. Moreover, the result represents an exciting technology with the strong potential to realize fully flexible consumer electronics such as application processor (AP) for mobile operating system, high-capacity memory, and wireless communication in the near future."
This result was published in the May online issue of the American Chemical Society"s journal, ACS Nano (In vivo Flexible RFICs Monolithically Encapsulated with LCP). They are currently engaged in commercializing efforts of roll-to-roll printing of flexible LSI on large area plastic substrates.
Movie at Youtube Link: Fabrication process for flexible LSI for flexible display, wearable computer and artificial retina for in vivo biomedical application
http://www.youtube.com/watch?v=5PpbM7m2PPs&feature=youtu.be
Applications of in Vivo Flexible Large Scale Integrated Circuits
Top: In vivo flexible large scale integrated circuits (LSI); Bottom: Schematic of roll-to-roll printing of flexible LSI on large area plastics.
2013.06.09 View 12240
A KAIST research team developed in vivo flexible large scale integrated circuits
Daejeon, Republic of Korea, May 6th, 2013–-A team led by Professor Keon Jae Lee from the Department of Materials Science and Engineering at KAIST has developed in vivo silicon-based flexible large scale integrated circuits (LSI) for bio-medical wireless communication.
Silicon-based semiconductors have played significant roles in signal processing, nerve stimulation, memory storage, and wireless communication in implantable electronics. However, the rigid and bulky LSI chips have limited uses in in vivo devices due to incongruent contact with the curvilinear surfaces of human organs. Especially, artificial retinas recently approved by the Food and Drug Administration (refer to the press release of FDA"s artificial retina approval) require extremely flexible and slim LSI to incorporate it within the cramped area of the human eye.
Although several research teams have fabricated flexible integrated circuits (ICs, tens of interconnected transistors) on plastics, their inaccurate nano-scale alignment on plastics has restricted the demonstration of flexible nano-transistors and their large scale interconnection for in vivo LSI applications such as main process unit (MPU), high density memory and wireless communication. Professor Lee"s team previously demonstrated fully functional flexible memory using ultrathin silicon membranes (Nano Letters, Flexible Memristive Memory Array on Plastic Substrates), however, its integration level and transistor size (over micron scale) have limited functional applications for flexible consumer electronics.
Professor Keon Jae Lee"s team fabricated radio frequency integrated circuits (RFICs) interconnected with thousand nano-transistors on silicon wafer by state-of-the-art CMOS process, and then they removed the entire bottom substrate except top 100 nm active circuit layer by wet chemical etching. The flexible RF switches for wireless communication were monolithically encapsulated with biocompatible liquid crystal polymers (LCPs) for in vivo bio-medical applications. Finally, they implanted the LCP encapsulated RFICs into live rats to demonstrate the stable operation of flexible devices under in vivo circumstances.
Professor Lee said, "This work could provide an approach to flexible LSI for an ideal artificial retina system and other bio-medical devices. Moreover, the result represents an exciting technology with the strong potential to realize fully flexible consumer electronics such as application processor (AP) for mobile operating system, high-capacity memory, and wireless communication in the near future."
This result was published in the May online issue of the American Chemical Society"s journal, ACS Nano (In vivo Flexible RFICs Monolithically Encapsulated with LCP). They are currently engaged in commercializing efforts of roll-to-roll printing of flexible LSI on large area plastic substrates.
Movie at Youtube Link: Fabrication process for flexible LSI for flexible display, wearable computer and artificial retina for in vivo biomedical application
http://www.youtube.com/watch?v=5PpbM7m2PPs&feature=youtu.be
Applications of in Vivo Flexible Large Scale Integrated Circuits
Top: In vivo flexible large scale integrated circuits (LSI); Bottom: Schematic of roll-to-roll printing of flexible LSI on large area plastics.
2013.06.09 View 12240 -
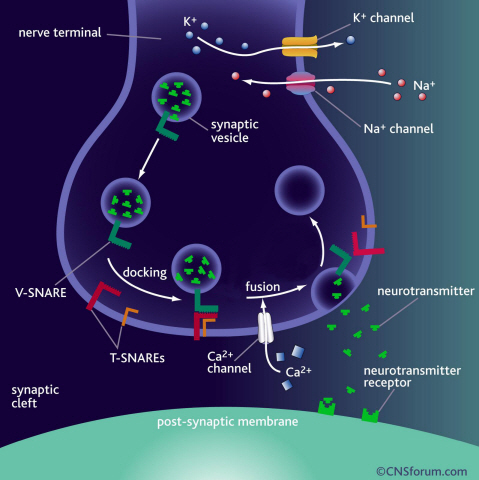 Neurotransmitter protein structure and operation principle identified
Professor Tae-Young Yoon
- Real-time measurement of structural change of bio-membrane fusion protein
- A new clue to degenerative brain diseases research
KAIST Physics Department’s Professor Tae-Young Yoon has successfully identified the hidden structure and operation mechanism of the SNARE protein, which has a central role in transporting neurotransmitters between neurons, using magnetic nanotweezers. SNARE protein’s cell membrane fusion function is closely related to degenerative brain diseases or neurological disorders such as Alzheimer’s. Hence, this research may provide a clue to the disease’s prevention and treatment.
Neurotransmission occurs when vesicles containing neurotransmitters fuse with cell membranes in neuron synapses. The SNARE protein is a cell-membrane fusion protein with a core role of releasing neurotransmitters. The academia speculated the SNARE protein would regulate the exchange of neurotransmitters, but its precise function and structure has been unknown. Professor Yoon’s research team developed an experimental technique using nanotweezers to measure physical changes to nanometer level by pulling and releasing each protein with force of 1 pN (piconewton). The research identified the existence of hidden SNARE protein"s intermediate structure. The process of withstanding and maintaining repulsive forces between bio-membranes in the hidden intermediate structure of SNARE to regulate the exchange of neurotransmitters has also been identified.
Professor Yoon’s research team developed an experimental technique using magnetic nanotweezers to measure physical changes of proteins to nanometer level by pulling and releasing each protein with force of 1 pN. The research identified the existence of hidden SNARE protein"s intermediate structure and its formation. The process of withstanding and maintaining repulsive forces between bio-membranes in the hidden intermediate structure of SNARE to regulate the exchange of neurotransmitters has also been discovered.
Professor Yoon said, “Ground breaking research results have been produced. A simple experimental technique of applying the smallest possible forces to proteins (with tweezers) to see their hidden structure and formation process can produce the same result as real observation has been developed.” He continued, “This technique will be very important in researching biological object with physical experimental technique. It will be a vital foundation to consilient research of different academia in the future.”
This research was a joint project of Physics Department’s Professor Tae-Young Yoon, KAIST, and Biomedical Engineering Institute’s Professor Yeon-Kyun Shin at KIST. KAIST Physics Department’s Professor Yong-Hoon Cho, Ph.D. candidate Do-Yong Lee and KIAS Computational Sciences Department’s Professor Chang-Bong Hyun participated. The research was published on Nature Communications on April 16th.
a) Neurotransmission occurs when vesicles containing neurotransmitters fuse with cell membranes in neuron synapses. A SNARE protein is a cell-membrane fusion protein with a core role of releasing neurotransmitters.
b) A schematic diagram using magnetic nanotweezers to measure protein structure changes on molecular level. The nanotweezers exert an exquisite pull and release of each protein with a force of 1 pN to measure physical changes to nanometer level in real-time to observe the hidden intermediate structure and operation principles of bio-membrane fusion protein.
2013.05.25 View 8514
Neurotransmitter protein structure and operation principle identified
Professor Tae-Young Yoon
- Real-time measurement of structural change of bio-membrane fusion protein
- A new clue to degenerative brain diseases research
KAIST Physics Department’s Professor Tae-Young Yoon has successfully identified the hidden structure and operation mechanism of the SNARE protein, which has a central role in transporting neurotransmitters between neurons, using magnetic nanotweezers. SNARE protein’s cell membrane fusion function is closely related to degenerative brain diseases or neurological disorders such as Alzheimer’s. Hence, this research may provide a clue to the disease’s prevention and treatment.
Neurotransmission occurs when vesicles containing neurotransmitters fuse with cell membranes in neuron synapses. The SNARE protein is a cell-membrane fusion protein with a core role of releasing neurotransmitters. The academia speculated the SNARE protein would regulate the exchange of neurotransmitters, but its precise function and structure has been unknown. Professor Yoon’s research team developed an experimental technique using nanotweezers to measure physical changes to nanometer level by pulling and releasing each protein with force of 1 pN (piconewton). The research identified the existence of hidden SNARE protein"s intermediate structure. The process of withstanding and maintaining repulsive forces between bio-membranes in the hidden intermediate structure of SNARE to regulate the exchange of neurotransmitters has also been identified.
Professor Yoon’s research team developed an experimental technique using magnetic nanotweezers to measure physical changes of proteins to nanometer level by pulling and releasing each protein with force of 1 pN. The research identified the existence of hidden SNARE protein"s intermediate structure and its formation. The process of withstanding and maintaining repulsive forces between bio-membranes in the hidden intermediate structure of SNARE to regulate the exchange of neurotransmitters has also been discovered.
Professor Yoon said, “Ground breaking research results have been produced. A simple experimental technique of applying the smallest possible forces to proteins (with tweezers) to see their hidden structure and formation process can produce the same result as real observation has been developed.” He continued, “This technique will be very important in researching biological object with physical experimental technique. It will be a vital foundation to consilient research of different academia in the future.”
This research was a joint project of Physics Department’s Professor Tae-Young Yoon, KAIST, and Biomedical Engineering Institute’s Professor Yeon-Kyun Shin at KIST. KAIST Physics Department’s Professor Yong-Hoon Cho, Ph.D. candidate Do-Yong Lee and KIAS Computational Sciences Department’s Professor Chang-Bong Hyun participated. The research was published on Nature Communications on April 16th.
a) Neurotransmission occurs when vesicles containing neurotransmitters fuse with cell membranes in neuron synapses. A SNARE protein is a cell-membrane fusion protein with a core role of releasing neurotransmitters.
b) A schematic diagram using magnetic nanotweezers to measure protein structure changes on molecular level. The nanotweezers exert an exquisite pull and release of each protein with a force of 1 pN to measure physical changes to nanometer level in real-time to observe the hidden intermediate structure and operation principles of bio-membrane fusion protein.
2013.05.25 View 8514 -
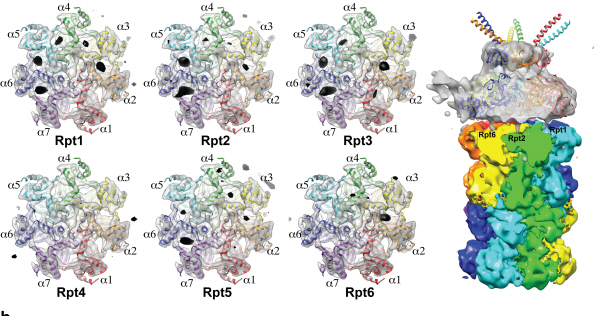 Complex responsible for protein breakdown in cells identified using Bio TEM
Professor Ho-Min Kim
- High resolution 3D structure analysis success using Bio Transmission Electron Microscopy (TEM), a giant step towards new anticancer treatment development
- Published in Nature on May 5th
Using TEM to observe protein molecules and analysing its high resolution 3D structure is now possible. KAIST Biomedical Science and Engineering Department’s Professor Ho-Min Kim has identified the high resolution structure of proteasome complexes, which is responsible for protein breakdown in cells, using Bio TEM.
This research has been published on the world"s most prestigious journal, Nature, online on May 5th. Our body controls many cellular processes through production and degradation of proteins to maintain homeostasis. A proteasome complex acts as a garbage disposal system and degrades cellular proteins when needed for regulation, which is one of the central roles of the body.
However, a mutation in proteasome complex leads to diseases such as cancer, degenerative brain diseases, and autoimmune diseases.
Currently, the anticancer drug Velcade is used to decrease proteasome function to treat Multiple Myeloma, a form of blood cancer. Research concerning proteasome complexes for more effective anticancer drugs and treatments with fewer side effects has been taking place for more than 20 years. There have been many difficulties in understanding proteasome function through 3D structure analysis since a proteasome complex, consisting of around 30 different proteins, has a great size and complexity.
The research team used Bio TEM instead of conventionally used protein crystallography technique. The protein sample was inserted into Bio TEM, hundreds of photographs were taken from various angles, and then a high–performance computer was used to analyse its structure. Bio TEM requires a smaller sample and can analyse the complexes of great size of proteins.
Professor Ho-Min Kim said, “Identifying proteasome complex assembly process and 3D structure will increase our understanding of cellular protein degradation process and hence assist in new drug development using this knowledge.” He added, “High resolution protein structure analysis using Bio TEM, used for the first time in Korea, will enable us to observe structure analysis of large protein complexes that were difficult to approach using protein crystallography.” Professor Kim continued, “If protein crystallography technology and Bio TEM could be used together to complement one another, it would bring a great synergetic effect to protein complex 3D structure analysis research in the future.”
Professor Ho-Min Kim has conducted this research since his post-doctorate at the University of California, San Francisco, under the advice of Professor Yifan Cheng; in co-operation with Harvard University and Colorado University.
Figure 1: A picture taken by Bio TEM of open state protein sample (proteasome complex)
Figure 2: Bio TEM image analysis showing protein 3D structure
2013.05.25 View 10100
Complex responsible for protein breakdown in cells identified using Bio TEM
Professor Ho-Min Kim
- High resolution 3D structure analysis success using Bio Transmission Electron Microscopy (TEM), a giant step towards new anticancer treatment development
- Published in Nature on May 5th
Using TEM to observe protein molecules and analysing its high resolution 3D structure is now possible. KAIST Biomedical Science and Engineering Department’s Professor Ho-Min Kim has identified the high resolution structure of proteasome complexes, which is responsible for protein breakdown in cells, using Bio TEM.
This research has been published on the world"s most prestigious journal, Nature, online on May 5th. Our body controls many cellular processes through production and degradation of proteins to maintain homeostasis. A proteasome complex acts as a garbage disposal system and degrades cellular proteins when needed for regulation, which is one of the central roles of the body.
However, a mutation in proteasome complex leads to diseases such as cancer, degenerative brain diseases, and autoimmune diseases.
Currently, the anticancer drug Velcade is used to decrease proteasome function to treat Multiple Myeloma, a form of blood cancer. Research concerning proteasome complexes for more effective anticancer drugs and treatments with fewer side effects has been taking place for more than 20 years. There have been many difficulties in understanding proteasome function through 3D structure analysis since a proteasome complex, consisting of around 30 different proteins, has a great size and complexity.
The research team used Bio TEM instead of conventionally used protein crystallography technique. The protein sample was inserted into Bio TEM, hundreds of photographs were taken from various angles, and then a high–performance computer was used to analyse its structure. Bio TEM requires a smaller sample and can analyse the complexes of great size of proteins.
Professor Ho-Min Kim said, “Identifying proteasome complex assembly process and 3D structure will increase our understanding of cellular protein degradation process and hence assist in new drug development using this knowledge.” He added, “High resolution protein structure analysis using Bio TEM, used for the first time in Korea, will enable us to observe structure analysis of large protein complexes that were difficult to approach using protein crystallography.” Professor Kim continued, “If protein crystallography technology and Bio TEM could be used together to complement one another, it would bring a great synergetic effect to protein complex 3D structure analysis research in the future.”
Professor Ho-Min Kim has conducted this research since his post-doctorate at the University of California, San Francisco, under the advice of Professor Yifan Cheng; in co-operation with Harvard University and Colorado University.
Figure 1: A picture taken by Bio TEM of open state protein sample (proteasome complex)
Figure 2: Bio TEM image analysis showing protein 3D structure
2013.05.25 View 10100 -
 KAIST develops a low-power 60 GHz radio frequency chip for mobile devices
As the capacity of handheld devices increases to accommodate a greater number of functions, these devices have more memory, larger display screens, and the ability to play higher definition video files. If the users of mobile devices, including smartphones, tablet PCs, and notebooks, want to share or transfer data on one device with that of another device, a great deal of time and effort are needed.
As a possible method for the speedy transmission of large data, researchers are studying the adoption of gigabits per second (Gbps) wireless communications operating over the 60 gigahertz (GHz) frequency band. Some commercial approaches have been introduced for full-HD video streaming from a fixed source to a display by using the 60 GHz band. But mobile applications have not been developed yet because the 60 GHz radio frequency (RF) circuit consumes hundreds of milliwatts (mW) of DC power.
Professor Chul Soon Park from the Department of Electrical Engineering at the Korea Advanced Institute of Science and Technology (KAIST) and his research team recently developed a low-power version of the 60 GHz radio frequency integrated circuit (RFIC). Inside the circuit are an energy-efficient modulator performing amplification as well as modulation and a sensitivity-improved receiver employing a gain boosting demodulator.
The research team said that their RFIC draws as little as 67 mW of power in the 60 GHz frequency band, consuming 31mW to send and 36mW to receive large volumes of data. RFIC is also small enough to be mounted on smartphones or notebooks, requiring only one chip (its width, length, and height are about 1 mm) and one antenna (4x5x1 mm3) for sending and receiving data with an integrated switch.
Professor Park, Director of the Intelligent Radio Engineering Center at KAIST, gave an upbeat assessment of the potential of RFIC for future applications. What we have developed is a low-power 60-GHz RF chip with a transmission speed of 10.7 gigabits per second. In tests, we were able to stream uncompressed full-HD videos from a smartphone or notebook to a display without a cable connection (Youtube Link: http://www.youtube.com/watch?v=6PVSLBhMymc). Our chip can be installed on mobile devices or even on cameras so that the devices are virtually connected to other devices and able to exchange large data with each other."
2013.04.02 View 8038
KAIST develops a low-power 60 GHz radio frequency chip for mobile devices
As the capacity of handheld devices increases to accommodate a greater number of functions, these devices have more memory, larger display screens, and the ability to play higher definition video files. If the users of mobile devices, including smartphones, tablet PCs, and notebooks, want to share or transfer data on one device with that of another device, a great deal of time and effort are needed.
As a possible method for the speedy transmission of large data, researchers are studying the adoption of gigabits per second (Gbps) wireless communications operating over the 60 gigahertz (GHz) frequency band. Some commercial approaches have been introduced for full-HD video streaming from a fixed source to a display by using the 60 GHz band. But mobile applications have not been developed yet because the 60 GHz radio frequency (RF) circuit consumes hundreds of milliwatts (mW) of DC power.
Professor Chul Soon Park from the Department of Electrical Engineering at the Korea Advanced Institute of Science and Technology (KAIST) and his research team recently developed a low-power version of the 60 GHz radio frequency integrated circuit (RFIC). Inside the circuit are an energy-efficient modulator performing amplification as well as modulation and a sensitivity-improved receiver employing a gain boosting demodulator.
The research team said that their RFIC draws as little as 67 mW of power in the 60 GHz frequency band, consuming 31mW to send and 36mW to receive large volumes of data. RFIC is also small enough to be mounted on smartphones or notebooks, requiring only one chip (its width, length, and height are about 1 mm) and one antenna (4x5x1 mm3) for sending and receiving data with an integrated switch.
Professor Park, Director of the Intelligent Radio Engineering Center at KAIST, gave an upbeat assessment of the potential of RFIC for future applications. What we have developed is a low-power 60-GHz RF chip with a transmission speed of 10.7 gigabits per second. In tests, we were able to stream uncompressed full-HD videos from a smartphone or notebook to a display without a cable connection (Youtube Link: http://www.youtube.com/watch?v=6PVSLBhMymc). Our chip can be installed on mobile devices or even on cameras so that the devices are virtually connected to other devices and able to exchange large data with each other."
2013.04.02 View 8038 -
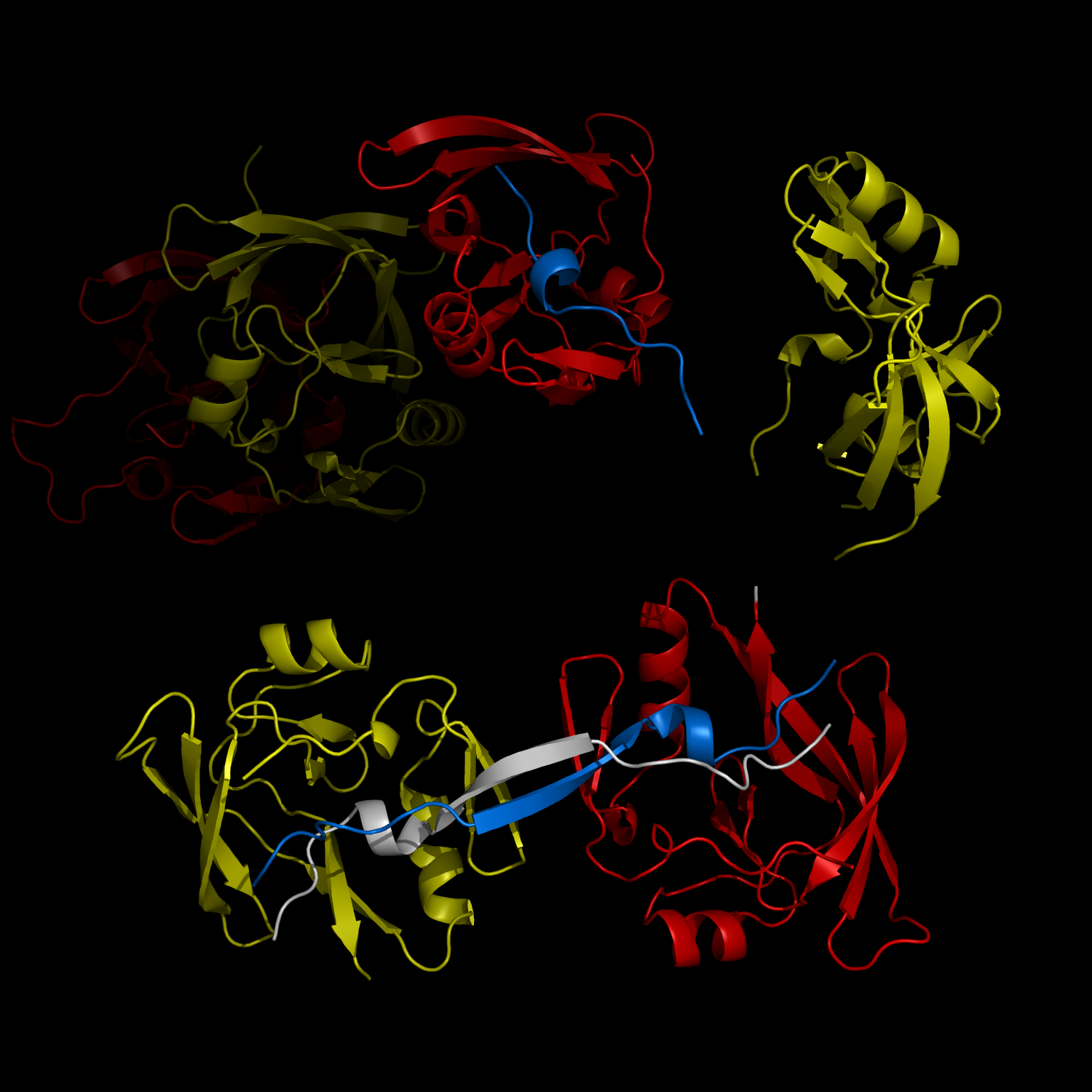 New Structural Insight into Neurodegenerative Disease
A research team from the Korea Advanced Institute of Science and Technology (KAIST) released their results on the structure and molecular details of the neurodegenerative disease-associated protein Ataxin-1. Mutations in Ataxin-1 cause the neurological disease, Spinocerebella Ataxia Type 1 (SCA1), which is characterized by a loss of muscular coordination and balance (ataxia), as is seen in Parkinson’s, Alzheimer’s, and Huntington’s diseases.
SCA1-causing mutations in the ATAXIN1 gene alter the length of a glutamine stretch in the Ataxin-1 protein. The research team provides the first structural insight into the complex formation of ATAXIN-1 with its binding partner, Capicua (CIC). The team, led by Professor Ji-Joon Song from the Department of Biological Sciences at KAIST, solved the structure of Ataxin-1 and CIC complex in atomic level revealing molecular details of the interaction between Ataxin-1 and CIC.
Professor Song explained his recent research work,
“We are able to see the intricate process of complex formation and reconfiguration of the two proteins when they interact with each other. Our work, we expect, will provide a new therapeutic target to modulate SCA1 neurodegenerative disease.”
Understanding structural and molecular details of proteins at the atomic level will help researchers to track the molecular pathogenesis of the disease and, ultimately, design targeted therapies or treatments for patients, rather than just relieving the symptoms of diseases.
Professor Song’s research paper, entitled “Structural Basis of Protein Complex Formation and Reconfiguration by Polyglutamine Disease Protein ATAXIN-1 and Capicua,” will be published in the March 15th issue of Genes & Development (www.genesdev.org).
Complex Formation and Reconfiguration of ATAXIN-1 and Capicua
The complex formation between a polyglutamine disease protein, ATXIN-1 and the transcriptional repressor Capicua (CIC) plays a critical role in SCA 1 pathogenesis. The image shows that the homodimerization of ATXIN-1 (yellow and red) is disrupted upon binding of CIC (blue). Furthermore, the binding of CIC to the ATXIN-1 induces a new form of ATXIN-1 dimerization mediated by CICs (ATXIN-1 AXH domains are shown in yellow and red, and CIC peptides shown in blue and white).
2013.04.02 View 8433
New Structural Insight into Neurodegenerative Disease
A research team from the Korea Advanced Institute of Science and Technology (KAIST) released their results on the structure and molecular details of the neurodegenerative disease-associated protein Ataxin-1. Mutations in Ataxin-1 cause the neurological disease, Spinocerebella Ataxia Type 1 (SCA1), which is characterized by a loss of muscular coordination and balance (ataxia), as is seen in Parkinson’s, Alzheimer’s, and Huntington’s diseases.
SCA1-causing mutations in the ATAXIN1 gene alter the length of a glutamine stretch in the Ataxin-1 protein. The research team provides the first structural insight into the complex formation of ATAXIN-1 with its binding partner, Capicua (CIC). The team, led by Professor Ji-Joon Song from the Department of Biological Sciences at KAIST, solved the structure of Ataxin-1 and CIC complex in atomic level revealing molecular details of the interaction between Ataxin-1 and CIC.
Professor Song explained his recent research work,
“We are able to see the intricate process of complex formation and reconfiguration of the two proteins when they interact with each other. Our work, we expect, will provide a new therapeutic target to modulate SCA1 neurodegenerative disease.”
Understanding structural and molecular details of proteins at the atomic level will help researchers to track the molecular pathogenesis of the disease and, ultimately, design targeted therapies or treatments for patients, rather than just relieving the symptoms of diseases.
Professor Song’s research paper, entitled “Structural Basis of Protein Complex Formation and Reconfiguration by Polyglutamine Disease Protein ATAXIN-1 and Capicua,” will be published in the March 15th issue of Genes & Development (www.genesdev.org).
Complex Formation and Reconfiguration of ATAXIN-1 and Capicua
The complex formation between a polyglutamine disease protein, ATXIN-1 and the transcriptional repressor Capicua (CIC) plays a critical role in SCA 1 pathogenesis. The image shows that the homodimerization of ATXIN-1 (yellow and red) is disrupted upon binding of CIC (blue). Furthermore, the binding of CIC to the ATXIN-1 induces a new form of ATXIN-1 dimerization mediated by CICs (ATXIN-1 AXH domains are shown in yellow and red, and CIC peptides shown in blue and white).
2013.04.02 View 8433 -
 The new era of personalized cancer diagnosis and treatment
Professor Tae-Young Yoon
- Succeeded in observing carcinogenic protein at the molecular level
- “Paved the way to customized cancer treatment through accurate analysis of carcinogenic protein”
The joint KAIST research team of Professor Tae Young Yoon of the Department of Physics and Professor Won Do Huh of the Department of Biological Sciences have developed the technology to monitor characteristics of carcinogenic protein in cancer tissue – for the first time in the world.
The technology makes it possible to analyse the mechanism of cancer development through a small amount of carcinogenic protein from a cancer patient. Therefore, a personalised approach to diagnosis and treatment using the knowledge of the specific mechanism of cancer development in the patient may be possible in the future.
Until recently, modern medicine could only speculate on the cause of cancer through statistics. Although developed countries, such as the United States, are known to use a large sequencing technology that analyses the patient’s DNA, identification of the interactions between proteins responsible for causing cancer remained an unanswered question for a long time in medicine.
Firstly, Professor Yoon’s research team has developed a fluorescent microscope that can observe even a single molecule. Then, the “Immunoprecipitation method”, a technology to extract a specific protein exploiting the high affinity between antigens and antibodies was developed. Using this technology and the microscope, “Real-Time Single Molecule co-Immunoprecipitation Method” was created. In this way, the team succeeded in observing the interactions between carcinogenic and other proteins at a molecular level, in real time.
To validate the developed technology, the team investigated Ras, a carcinogenic protein; its mutation statistically is known to cause around 30% of cancers.
The experimental results confirmed that 30-50% of Ras protein was expressed in mouse tumour and human cancer cells. In normal cells, less than 5% of Ras protein was expressed. Thus, the experiment showed that unusual increase in activation of Ras protein induces cancer.
The increase in the ratio of active Ras protein can be inferred from existing research data but the measurement of specific numerical data has never been done before.
The team suggested a new molecular level diagnosis technique of identifying the progress of cancer in patients through measuring the percentage of activated carcinogenic protein in cancer tissue.
Professor Yoon Tae-young said, “This newly developed technology does not require a separate procedure of protein expression or refining, hence the existing proteins in real biological tissues or cancer cells can be observed directly.” He also said, “Since carcinogenic protein can be analyzed accurately, it has opened up the path to customized cancer treatment in the future.”
“Since the observation is possible on a molecular level, the technology confers the advantage that researchers can carry out various examinations on a small sample of the cancer patient.” He added, “The clinical trial will start in December 2012 and in a few years customized cancer diagnosis and treatment will be possible.”
Meanwhile, the research has been published in Nature Communications (February 19). Many researchers from various fields have participated, regardless of the differences in their speciality, and successfully produced interdisciplinary research. Professor Tae Young Yoon of the Department of Physics and Professors Dae Sik Lim and Won Do Huh of Biological Sciences at KAIST, and Professor Chang Bong Hyun of Computational Science of KIAS contributed to developing the technique.
Figure 1: Schematic diagram of observed interactions at the molecular level in real time using fluorescent microscope. The carcinogenic protein from a mouse tumour is fixed on the microchip, and its molecular characteristics are observed live.
Figure 2: Molecular interaction data using a molecular level fluorescent microscope. A signal in the form of spike is shown when two proteins combine. This is monitored live using an Electron Multiplying Charge Coupled Device (EMCCD). It shows signal results in bright dots.
An organism has an immune system as a defence mechanism to foreign intruders. The immune system is activated when unwanted pathogens or foreign protein are in the body. Antibodies form in recognition of the specific antigen to protect itself. Organisms evolved to form antibodies with high specificity to a certain antigen. Antibodies only react to its complementary antigens. The field of molecular biology uses the affinity between antigens and antibodies to extract specific proteins; a technology called immunoprecipitation. Even in a mixture of many proteins, the protein sought can be extracted using antibodies. Thus immunoprecipitation is widely used to detect pathogens or to extract specific proteins.
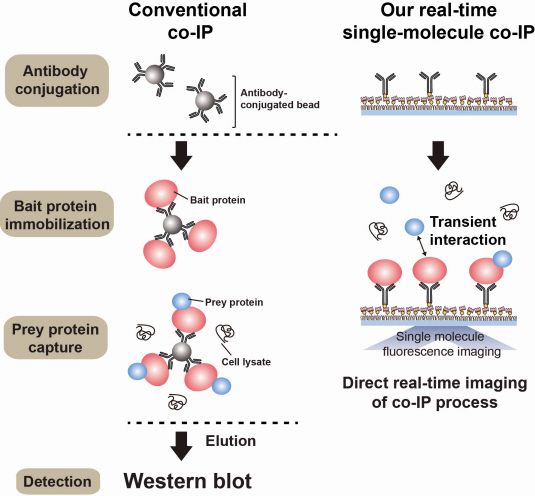
Technology co-IP is a well-known example that uses immunoprecipitation. The research on interactions between proteins uses co-IP in general. The basis of fixing the antigen on the antibody to extract antigen protein is the same as immunoprecipitation. Then, researchers inject and observe its reaction with the partner protein to observe the interactions and precipitate the antibodies. If the reaction occurs, the partner protein will be found with the antibodies in the precipitations. If not, then the partner protein will not be found. This shows that the two proteins interact.
However, the traditional co-IP can be used to infer the interactions between the two proteins although the information of the dynamics on how the reaction occurs is lost. To overcome these shortcomings, the Real-Time Single Molecule co-IP Method enables observation on individual protein level in real time. Therefore, the significance of the new technique is in making observation of interactions more direct and quantitative.
Additional Figure 1: Comparison between Conventional co-IP and Real-Time Single Molecule co-IP
2013.04.01 View 16977
The new era of personalized cancer diagnosis and treatment
Professor Tae-Young Yoon
- Succeeded in observing carcinogenic protein at the molecular level
- “Paved the way to customized cancer treatment through accurate analysis of carcinogenic protein”
The joint KAIST research team of Professor Tae Young Yoon of the Department of Physics and Professor Won Do Huh of the Department of Biological Sciences have developed the technology to monitor characteristics of carcinogenic protein in cancer tissue – for the first time in the world.
The technology makes it possible to analyse the mechanism of cancer development through a small amount of carcinogenic protein from a cancer patient. Therefore, a personalised approach to diagnosis and treatment using the knowledge of the specific mechanism of cancer development in the patient may be possible in the future.
Until recently, modern medicine could only speculate on the cause of cancer through statistics. Although developed countries, such as the United States, are known to use a large sequencing technology that analyses the patient’s DNA, identification of the interactions between proteins responsible for causing cancer remained an unanswered question for a long time in medicine.
Firstly, Professor Yoon’s research team has developed a fluorescent microscope that can observe even a single molecule. Then, the “Immunoprecipitation method”, a technology to extract a specific protein exploiting the high affinity between antigens and antibodies was developed. Using this technology and the microscope, “Real-Time Single Molecule co-Immunoprecipitation Method” was created. In this way, the team succeeded in observing the interactions between carcinogenic and other proteins at a molecular level, in real time.
To validate the developed technology, the team investigated Ras, a carcinogenic protein; its mutation statistically is known to cause around 30% of cancers.
The experimental results confirmed that 30-50% of Ras protein was expressed in mouse tumour and human cancer cells. In normal cells, less than 5% of Ras protein was expressed. Thus, the experiment showed that unusual increase in activation of Ras protein induces cancer.
The increase in the ratio of active Ras protein can be inferred from existing research data but the measurement of specific numerical data has never been done before.
The team suggested a new molecular level diagnosis technique of identifying the progress of cancer in patients through measuring the percentage of activated carcinogenic protein in cancer tissue.
Professor Yoon Tae-young said, “This newly developed technology does not require a separate procedure of protein expression or refining, hence the existing proteins in real biological tissues or cancer cells can be observed directly.” He also said, “Since carcinogenic protein can be analyzed accurately, it has opened up the path to customized cancer treatment in the future.”
“Since the observation is possible on a molecular level, the technology confers the advantage that researchers can carry out various examinations on a small sample of the cancer patient.” He added, “The clinical trial will start in December 2012 and in a few years customized cancer diagnosis and treatment will be possible.”
Meanwhile, the research has been published in Nature Communications (February 19). Many researchers from various fields have participated, regardless of the differences in their speciality, and successfully produced interdisciplinary research. Professor Tae Young Yoon of the Department of Physics and Professors Dae Sik Lim and Won Do Huh of Biological Sciences at KAIST, and Professor Chang Bong Hyun of Computational Science of KIAS contributed to developing the technique.
Figure 1: Schematic diagram of observed interactions at the molecular level in real time using fluorescent microscope. The carcinogenic protein from a mouse tumour is fixed on the microchip, and its molecular characteristics are observed live.
Figure 2: Molecular interaction data using a molecular level fluorescent microscope. A signal in the form of spike is shown when two proteins combine. This is monitored live using an Electron Multiplying Charge Coupled Device (EMCCD). It shows signal results in bright dots.
An organism has an immune system as a defence mechanism to foreign intruders. The immune system is activated when unwanted pathogens or foreign protein are in the body. Antibodies form in recognition of the specific antigen to protect itself. Organisms evolved to form antibodies with high specificity to a certain antigen. Antibodies only react to its complementary antigens. The field of molecular biology uses the affinity between antigens and antibodies to extract specific proteins; a technology called immunoprecipitation. Even in a mixture of many proteins, the protein sought can be extracted using antibodies. Thus immunoprecipitation is widely used to detect pathogens or to extract specific proteins.
Technology co-IP is a well-known example that uses immunoprecipitation. The research on interactions between proteins uses co-IP in general. The basis of fixing the antigen on the antibody to extract antigen protein is the same as immunoprecipitation. Then, researchers inject and observe its reaction with the partner protein to observe the interactions and precipitate the antibodies. If the reaction occurs, the partner protein will be found with the antibodies in the precipitations. If not, then the partner protein will not be found. This shows that the two proteins interact.
However, the traditional co-IP can be used to infer the interactions between the two proteins although the information of the dynamics on how the reaction occurs is lost. To overcome these shortcomings, the Real-Time Single Molecule co-IP Method enables observation on individual protein level in real time. Therefore, the significance of the new technique is in making observation of interactions more direct and quantitative.
Additional Figure 1: Comparison between Conventional co-IP and Real-Time Single Molecule co-IP
2013.04.01 View 16977 -
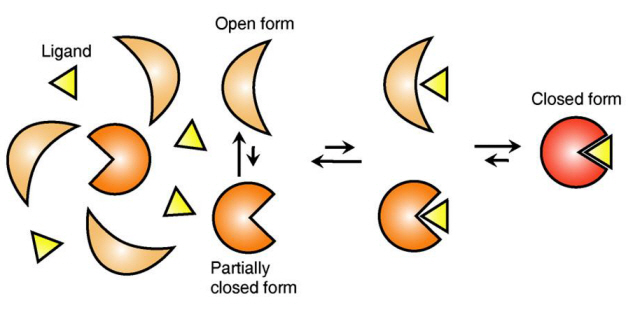 Ligand Recognition Mechanism of Protein Identified
Professor Hak-Sung Kim
-“Solved the 50 year old mystery of how protein recognises and binds to ligands”
- Exciting potential for understanding life phenomena and the further development of highly effective therapeutic agent development
KAIST’s Biological Science Department’s Professor Hak-Sung Kim, working in collaboration with Professor Sung-Chul Hong of Department of Physics, Seoul National University, has identified the mechanism of how the protein recognizes and binds to ligands within the human body.
The research findings were published in the online edition of Nature Chemical Biology (March 18), which is the most prestigious journal in the field of life science.
Since the research identified the mechanism, of which protein recognises and binds to ligands, it will take an essential role in understanding complex life phenomenon by understanding regulatory function of protein.
Also, ligand recognition of proteins is closely related to the cause of various diseases. Therefore the research team hopes to contribute to the development of highly effective treatments.
Ligands, well-known examples include nucleic acid and proteins, form the structure of an organism or are essential constituents with special functions such as information signalling.
In particular, the most important role of protein is recognising and binding to a particular ligand and hence regulating and maintaining life phenomena. The abnormal occurrence of an error in recognition of ligands may lead to various diseases.
The research team focused on the repetition of change in protein structure from the most stable “open form” to a relatively unstable “partially closed form”.
Professor Kim’s team analysed the change in protein structure when binding to a ligand on a molecular level in real time to explain the ligand recognition mechanism.
The research findings showed that ligands prefer the most stable protein structure. The team was the first in the world to identify that ligands alter protein structure to the most stable, the lowest energy level, when it binds to the protein.
In addition, the team found that ligands bind to unstable partially-closed forms to change protein structure.
The existing models to explain ligand recognition mechanism of protein are “Induced Custom Model”, which involves change in protein structure in binding to ligands, and the “Structure Selection Model”, which argues that ligands select and recognise only the best protein structure out of many. The academic world considers that the team’s research findings have perfectly proved the models through experiments for the first time in the world.
Professor Kim explained, “In the presence of ligands, there exists a phenomenon where the speed of altering protein structure is changed. This phenomenon is analysed on a molecular level to prove ligand recognition mechanism of protein for the first time”. He also said, “The 50-year old mystery, that existed only as a hypothesis on biology textbooks and was thought never to be solved, has been confirmed through experiments for the first time.”
Figure 1: Proteins, with open and partially open form, recognising and binding to ligands.
Figure 2: Ligands temporarily bind to a stable protein structure, open form, which changes into the most stable structure, closed form. In addition, binding to partially closed form also changes protein structure to closed form.
2013.04.01 View 10239
Ligand Recognition Mechanism of Protein Identified
Professor Hak-Sung Kim
-“Solved the 50 year old mystery of how protein recognises and binds to ligands”
- Exciting potential for understanding life phenomena and the further development of highly effective therapeutic agent development
KAIST’s Biological Science Department’s Professor Hak-Sung Kim, working in collaboration with Professor Sung-Chul Hong of Department of Physics, Seoul National University, has identified the mechanism of how the protein recognizes and binds to ligands within the human body.
The research findings were published in the online edition of Nature Chemical Biology (March 18), which is the most prestigious journal in the field of life science.
Since the research identified the mechanism, of which protein recognises and binds to ligands, it will take an essential role in understanding complex life phenomenon by understanding regulatory function of protein.
Also, ligand recognition of proteins is closely related to the cause of various diseases. Therefore the research team hopes to contribute to the development of highly effective treatments.
Ligands, well-known examples include nucleic acid and proteins, form the structure of an organism or are essential constituents with special functions such as information signalling.
In particular, the most important role of protein is recognising and binding to a particular ligand and hence regulating and maintaining life phenomena. The abnormal occurrence of an error in recognition of ligands may lead to various diseases.
The research team focused on the repetition of change in protein structure from the most stable “open form” to a relatively unstable “partially closed form”.
Professor Kim’s team analysed the change in protein structure when binding to a ligand on a molecular level in real time to explain the ligand recognition mechanism.
The research findings showed that ligands prefer the most stable protein structure. The team was the first in the world to identify that ligands alter protein structure to the most stable, the lowest energy level, when it binds to the protein.
In addition, the team found that ligands bind to unstable partially-closed forms to change protein structure.
The existing models to explain ligand recognition mechanism of protein are “Induced Custom Model”, which involves change in protein structure in binding to ligands, and the “Structure Selection Model”, which argues that ligands select and recognise only the best protein structure out of many. The academic world considers that the team’s research findings have perfectly proved the models through experiments for the first time in the world.
Professor Kim explained, “In the presence of ligands, there exists a phenomenon where the speed of altering protein structure is changed. This phenomenon is analysed on a molecular level to prove ligand recognition mechanism of protein for the first time”. He also said, “The 50-year old mystery, that existed only as a hypothesis on biology textbooks and was thought never to be solved, has been confirmed through experiments for the first time.”
Figure 1: Proteins, with open and partially open form, recognising and binding to ligands.
Figure 2: Ligands temporarily bind to a stable protein structure, open form, which changes into the most stable structure, closed form. In addition, binding to partially closed form also changes protein structure to closed form.
2013.04.01 View 10239