Department+of+Biological+Sciences
-
 Professor Won Do Heo on LED Light Technology for Controlling Proteins in Living Cells
With the newly developed LED technology, Professor Won Do Heo at the College of Life Science and Bioengineering, KAIST, was able to suppress cell migration and division when cells are exposed to LED light. This suggests a breakthrough to apply in future cancer cell research.
Professor Heo talked about the impact of his research in the following excerpt from a news article:
“We are already conducting research on the spread of cancer, as well as brain science in animal models with the Light-Activated Reversible Inhibition by Assembled Trap. I believe this technology will be a breakthrough in investigating cancer treatments and the function of neurons in a complex neural network, which existing technologies have not been able to do.”
From EE Times Europe, June 19, 2014
“LED Light Technology Controls Proteins in Living Cells”
http://www.ledlighting-eetimes.com/en/led-light-technology-controls-proteins-in-living-cells.html?cmp_id=7&news_id=222909336
2014.06.22 View 7475
Professor Won Do Heo on LED Light Technology for Controlling Proteins in Living Cells
With the newly developed LED technology, Professor Won Do Heo at the College of Life Science and Bioengineering, KAIST, was able to suppress cell migration and division when cells are exposed to LED light. This suggests a breakthrough to apply in future cancer cell research.
Professor Heo talked about the impact of his research in the following excerpt from a news article:
“We are already conducting research on the spread of cancer, as well as brain science in animal models with the Light-Activated Reversible Inhibition by Assembled Trap. I believe this technology will be a breakthrough in investigating cancer treatments and the function of neurons in a complex neural network, which existing technologies have not been able to do.”
From EE Times Europe, June 19, 2014
“LED Light Technology Controls Proteins in Living Cells”
http://www.ledlighting-eetimes.com/en/led-light-technology-controls-proteins-in-living-cells.html?cmp_id=7&news_id=222909336
2014.06.22 View 7475 -
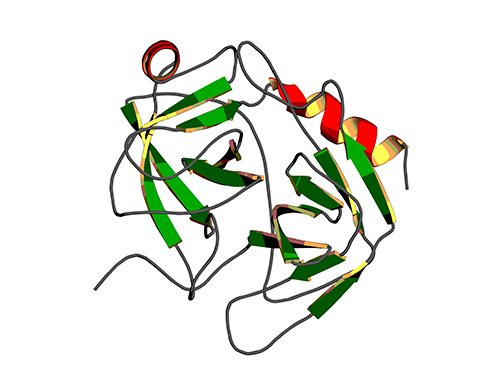 Binding Regulatory Mechanism of Protein Biomolecules Revealed
Professor Hak-Sung Kim
A research team led by Professor Hak-Sung Kim of Biological Sciences, KAIST, and Dr. Mun-Hyeong Seo, KAIST, has revealed a regulatory mechanism that controls the binding affinity of protein’s biomolecules, which is crucial for the protein to recognize molecules and carry out functions within the body.
The research results were published in the April 24th online edition of Nature Communications.
The protein, represented by enzyme, antibody, or hormones, specifically recognizes a variety of biomolecules in all organisms and implements signaling or immune response to precisely adjust and maintain important biological processes. The protein binding affinity of biomolecules plays a crucial role in determining the duration of the bond between two molecules, and hence to determine and control the in-vivo function of proteins.
The researchers have noted that, during the process of proteins’ recognizing biomolecules, the protein binding affinity of biomolecules is closely linked not only to the size of non-covalent interaction between two molecules, but also to the unique kinetic properties of proteins.
To identify the basic mechanism that determines the protein binding affinity of biomolecules, Professor Kim and his research team have made mutation in the allosteric site of protein to create a variety of mutant proteins with the same chemical binding surface, but with the binding affinity vastly differing from 10 to 100 times. The allosteric site of the protein refers to a region which does not directly bind with biomolecules, but crucially influences the biomolecule recognition site.
Using real-time analysis at the single-molecule level of unique kinetic properties of the produced mutant proteins, the researchers were able to identify that the protein binding affinity of biomolecules is directly associated with the protein’s specific kinetic characteristics, its structure opening rate.
Also, by proving that unique characteristics of the protein can be changed at the allosteric site, instead of protein’s direct binding site with biomolecules, the researchers have demonstrated a new methodology of regulating the in-vivo function of proteins.
The researchers expect that these results will contribute greatly to a deeper understanding of protein’s nature that governs various life phenomena and help evaluate the proof of interpreting protein binding affinity of biomolecules from the perspective of protein kinetics.
Professor Kim said, “Until now, the protein binding affinity of biomolecules was determined by a direct interaction between two molecules. Our research has identified an important fact that the structure opening rate of proteins also plays a crucial role in determining their binding affinity.”
[Picture]
A correlation graph of opening rate (kopening) and binding affinity (kd) between protein’s stable, open state and its unstable, partially closed state.
2014.05.02 View 9224
Binding Regulatory Mechanism of Protein Biomolecules Revealed
Professor Hak-Sung Kim
A research team led by Professor Hak-Sung Kim of Biological Sciences, KAIST, and Dr. Mun-Hyeong Seo, KAIST, has revealed a regulatory mechanism that controls the binding affinity of protein’s biomolecules, which is crucial for the protein to recognize molecules and carry out functions within the body.
The research results were published in the April 24th online edition of Nature Communications.
The protein, represented by enzyme, antibody, or hormones, specifically recognizes a variety of biomolecules in all organisms and implements signaling or immune response to precisely adjust and maintain important biological processes. The protein binding affinity of biomolecules plays a crucial role in determining the duration of the bond between two molecules, and hence to determine and control the in-vivo function of proteins.
The researchers have noted that, during the process of proteins’ recognizing biomolecules, the protein binding affinity of biomolecules is closely linked not only to the size of non-covalent interaction between two molecules, but also to the unique kinetic properties of proteins.
To identify the basic mechanism that determines the protein binding affinity of biomolecules, Professor Kim and his research team have made mutation in the allosteric site of protein to create a variety of mutant proteins with the same chemical binding surface, but with the binding affinity vastly differing from 10 to 100 times. The allosteric site of the protein refers to a region which does not directly bind with biomolecules, but crucially influences the biomolecule recognition site.
Using real-time analysis at the single-molecule level of unique kinetic properties of the produced mutant proteins, the researchers were able to identify that the protein binding affinity of biomolecules is directly associated with the protein’s specific kinetic characteristics, its structure opening rate.
Also, by proving that unique characteristics of the protein can be changed at the allosteric site, instead of protein’s direct binding site with biomolecules, the researchers have demonstrated a new methodology of regulating the in-vivo function of proteins.
The researchers expect that these results will contribute greatly to a deeper understanding of protein’s nature that governs various life phenomena and help evaluate the proof of interpreting protein binding affinity of biomolecules from the perspective of protein kinetics.
Professor Kim said, “Until now, the protein binding affinity of biomolecules was determined by a direct interaction between two molecules. Our research has identified an important fact that the structure opening rate of proteins also plays a crucial role in determining their binding affinity.”
[Picture]
A correlation graph of opening rate (kopening) and binding affinity (kd) between protein’s stable, open state and its unstable, partially closed state.
2014.05.02 View 9224 -
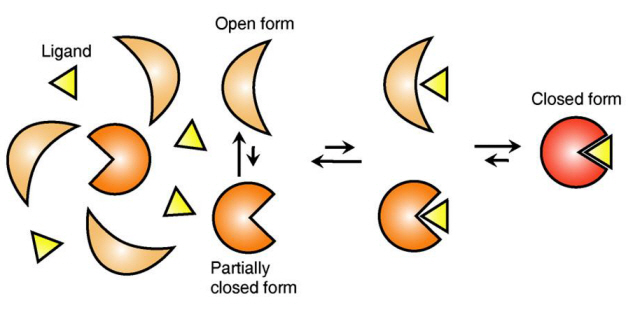 Ligand Recognition Mechanism of Protein Identified
Professor Hak-Sung Kim
-“Solved the 50 year old mystery of how protein recognises and binds to ligands”
- Exciting potential for understanding life phenomena and the further development of highly effective therapeutic agent development
KAIST’s Biological Science Department’s Professor Hak-Sung Kim, working in collaboration with Professor Sung-Chul Hong of Department of Physics, Seoul National University, has identified the mechanism of how the protein recognizes and binds to ligands within the human body.
The research findings were published in the online edition of Nature Chemical Biology (March 18), which is the most prestigious journal in the field of life science.
Since the research identified the mechanism, of which protein recognises and binds to ligands, it will take an essential role in understanding complex life phenomenon by understanding regulatory function of protein.
Also, ligand recognition of proteins is closely related to the cause of various diseases. Therefore the research team hopes to contribute to the development of highly effective treatments.
Ligands, well-known examples include nucleic acid and proteins, form the structure of an organism or are essential constituents with special functions such as information signalling.
In particular, the most important role of protein is recognising and binding to a particular ligand and hence regulating and maintaining life phenomena. The abnormal occurrence of an error in recognition of ligands may lead to various diseases.
The research team focused on the repetition of change in protein structure from the most stable “open form” to a relatively unstable “partially closed form”.
Professor Kim’s team analysed the change in protein structure when binding to a ligand on a molecular level in real time to explain the ligand recognition mechanism.
The research findings showed that ligands prefer the most stable protein structure. The team was the first in the world to identify that ligands alter protein structure to the most stable, the lowest energy level, when it binds to the protein.
In addition, the team found that ligands bind to unstable partially-closed forms to change protein structure.
The existing models to explain ligand recognition mechanism of protein are “Induced Custom Model”, which involves change in protein structure in binding to ligands, and the “Structure Selection Model”, which argues that ligands select and recognise only the best protein structure out of many. The academic world considers that the team’s research findings have perfectly proved the models through experiments for the first time in the world.
Professor Kim explained, “In the presence of ligands, there exists a phenomenon where the speed of altering protein structure is changed. This phenomenon is analysed on a molecular level to prove ligand recognition mechanism of protein for the first time”. He also said, “The 50-year old mystery, that existed only as a hypothesis on biology textbooks and was thought never to be solved, has been confirmed through experiments for the first time.”
Figure 1: Proteins, with open and partially open form, recognising and binding to ligands.
Figure 2: Ligands temporarily bind to a stable protein structure, open form, which changes into the most stable structure, closed form. In addition, binding to partially closed form also changes protein structure to closed form.
2013.04.01 View 10239
Ligand Recognition Mechanism of Protein Identified
Professor Hak-Sung Kim
-“Solved the 50 year old mystery of how protein recognises and binds to ligands”
- Exciting potential for understanding life phenomena and the further development of highly effective therapeutic agent development
KAIST’s Biological Science Department’s Professor Hak-Sung Kim, working in collaboration with Professor Sung-Chul Hong of Department of Physics, Seoul National University, has identified the mechanism of how the protein recognizes and binds to ligands within the human body.
The research findings were published in the online edition of Nature Chemical Biology (March 18), which is the most prestigious journal in the field of life science.
Since the research identified the mechanism, of which protein recognises and binds to ligands, it will take an essential role in understanding complex life phenomenon by understanding regulatory function of protein.
Also, ligand recognition of proteins is closely related to the cause of various diseases. Therefore the research team hopes to contribute to the development of highly effective treatments.
Ligands, well-known examples include nucleic acid and proteins, form the structure of an organism or are essential constituents with special functions such as information signalling.
In particular, the most important role of protein is recognising and binding to a particular ligand and hence regulating and maintaining life phenomena. The abnormal occurrence of an error in recognition of ligands may lead to various diseases.
The research team focused on the repetition of change in protein structure from the most stable “open form” to a relatively unstable “partially closed form”.
Professor Kim’s team analysed the change in protein structure when binding to a ligand on a molecular level in real time to explain the ligand recognition mechanism.
The research findings showed that ligands prefer the most stable protein structure. The team was the first in the world to identify that ligands alter protein structure to the most stable, the lowest energy level, when it binds to the protein.
In addition, the team found that ligands bind to unstable partially-closed forms to change protein structure.
The existing models to explain ligand recognition mechanism of protein are “Induced Custom Model”, which involves change in protein structure in binding to ligands, and the “Structure Selection Model”, which argues that ligands select and recognise only the best protein structure out of many. The academic world considers that the team’s research findings have perfectly proved the models through experiments for the first time in the world.
Professor Kim explained, “In the presence of ligands, there exists a phenomenon where the speed of altering protein structure is changed. This phenomenon is analysed on a molecular level to prove ligand recognition mechanism of protein for the first time”. He also said, “The 50-year old mystery, that existed only as a hypothesis on biology textbooks and was thought never to be solved, has been confirmed through experiments for the first time.”
Figure 1: Proteins, with open and partially open form, recognising and binding to ligands.
Figure 2: Ligands temporarily bind to a stable protein structure, open form, which changes into the most stable structure, closed form. In addition, binding to partially closed form also changes protein structure to closed form.
2013.04.01 View 10239 -
 The hereditary factor of autism revealed
Korean researchers have successfully investigated the causes and hereditary factors for autistic behavior and proposed a new treatment method with fewer side effects.
This research was jointly supported by the Ministry of Education, Science and Technology and the National Research Foundation as part of the Leading Researcher and Science Research Center Program
The research findings were publishing in the June edition of Nature magazine and will also be introduced in the July edition of Nature Reviews Drug Discovery, under the title ‘Autistic-like social behavior in Shank2-mutant mice improved by restoring NMDA receptor function’.
The research team found that lack of Shank2 genes in mice, which are responsible for the production of synapse proteins, caused autistic-like behavior. The results strongly suggested that the Shank2 gene was linked to autistic behavior and that Shank2 deficiency induced autistic behaviors.
Autism is a neural development disorder characterized by impaired social interaction, repetitive behavior, mental retardation, anxiety and hyperactivity. Around 100 million people worldwide display symptoms of autistic behavior. Recent studies conducted by the University of Washington revealed that 1 out of 3 young adults who display autistic behavior do not fit into the workplace or get accepted to college, a much higher rate than any other disorder. However, an effective cure has not yet been developed and current treatments are limited to reducing repetitive behavior.
The research team confirmed autistic-like social behavior in mice without the Shank2 genes and that the mice had decreased levels of neurotransmission in the NMDA receptor. The mice also showed damaged synaptic plasticity* in the hippocampus**.
* Plasticity: ability of the connectionbetween two neurons to change in strength in response to transmission of information
**Hippocampus: part of the brain responsible for short-term and long-term memory as well as spatial navigation.
The research team also found out that, to restore the function of the NMDA receptor, the passive stimulation of certain receptors, such as the mGLuR5, yielded better treatment results than the direct stimulation of the NMDA. This greatly reduces the side effects associated with the direct stimulation of receptors, resulting in a more effective treatment method.
This research successfully investigated the function of the Shank2 gene in the nerve tissue and showed how the reduced function of the NMDA receptor, due to the lack of the gene, resulted in autistic behavior. It also provided new possibilities for the treatment of autistic behavior and impaired social interaction
2012.06.24 View 10654
The hereditary factor of autism revealed
Korean researchers have successfully investigated the causes and hereditary factors for autistic behavior and proposed a new treatment method with fewer side effects.
This research was jointly supported by the Ministry of Education, Science and Technology and the National Research Foundation as part of the Leading Researcher and Science Research Center Program
The research findings were publishing in the June edition of Nature magazine and will also be introduced in the July edition of Nature Reviews Drug Discovery, under the title ‘Autistic-like social behavior in Shank2-mutant mice improved by restoring NMDA receptor function’.
The research team found that lack of Shank2 genes in mice, which are responsible for the production of synapse proteins, caused autistic-like behavior. The results strongly suggested that the Shank2 gene was linked to autistic behavior and that Shank2 deficiency induced autistic behaviors.
Autism is a neural development disorder characterized by impaired social interaction, repetitive behavior, mental retardation, anxiety and hyperactivity. Around 100 million people worldwide display symptoms of autistic behavior. Recent studies conducted by the University of Washington revealed that 1 out of 3 young adults who display autistic behavior do not fit into the workplace or get accepted to college, a much higher rate than any other disorder. However, an effective cure has not yet been developed and current treatments are limited to reducing repetitive behavior.
The research team confirmed autistic-like social behavior in mice without the Shank2 genes and that the mice had decreased levels of neurotransmission in the NMDA receptor. The mice also showed damaged synaptic plasticity* in the hippocampus**.
* Plasticity: ability of the connectionbetween two neurons to change in strength in response to transmission of information
**Hippocampus: part of the brain responsible for short-term and long-term memory as well as spatial navigation.
The research team also found out that, to restore the function of the NMDA receptor, the passive stimulation of certain receptors, such as the mGLuR5, yielded better treatment results than the direct stimulation of the NMDA. This greatly reduces the side effects associated with the direct stimulation of receptors, resulting in a more effective treatment method.
This research successfully investigated the function of the Shank2 gene in the nerve tissue and showed how the reduced function of the NMDA receptor, due to the lack of the gene, resulted in autistic behavior. It also provided new possibilities for the treatment of autistic behavior and impaired social interaction
2012.06.24 View 10654 -
 Successful Development of Excavation System of Biomarkers containing Protein Decomposition Control Enzyme Information
A Korean team of researchers successfully developed a biomarker excavation system named E3Net that excavates biomarkers containing information of the enzymes that control the decomposition of proteins. The development of the system paved the possibility of development of new high quality biomarkers.
*Biomarker: Molecular information of unique patterns derived from genes and proteins that allow the monitoring of physical changes from genetic or environmental causes.
Professor Lee Kwan Soo’s team (Department of Biological Sciences) composed of Doctorate candidate Han Young Woong, Lee Ho Dong Ph.D. and Professor Park Jong Chul published a dissertation in the April edition of Molecular and Cellular Proteomics. (Dissertation Title: A system for exploring E3-mediated regulatory networks of cellular functions).
Professor Lee’s team compiled all available information of the enzyme that controls protein decomposition (E3 enzyme) and successfully compiled the inter-substrate network by extracting information from 20,000 biology related data base dissertations. The result was the development of the E3Net system that analyzes the related cell function and disease.
Cells have a system that produces, destroys, and recycles proteins in response to the ever changing environmental conditions. Error in these processes leads to disease.
Therefore finding the relationship between E3 enzymes that control the decomposition of proteins and the substrates will allow disease curing and prevention to become much easier. E3 enzyme is responsible for 80% of the protein decomposition and is therefore predicted to be related to various diseases.
However the information on E3 enzyme and inter-substrate behavior are spread out among numerous dissertations and data bases which prevented methodological analysis of the role of the related cells and characteristics of the disease itself.
Professor Lee’s team was successful in creating the E3Net that compiled 2,201 pieces of E3 substrate information, 4,896 pieces of substrate information, and 1,671 pieces of inter-substrate relationship information. This compilation allows for the systematic analysis of cells and diseases.
The newly created network is 10 times larger than the existing network and is the first case where it is possible to accurately find the cell function and related diseases.
It is anticipated that the use of the E3Net will allow the excavation of new biomarkers for the development of personalized drug systems.
The research team applied the E3Net to find tens of new candidate biomarkers related to the major modern diseases like diabetes and cancer.
2012.05.30 View 11620
Successful Development of Excavation System of Biomarkers containing Protein Decomposition Control Enzyme Information
A Korean team of researchers successfully developed a biomarker excavation system named E3Net that excavates biomarkers containing information of the enzymes that control the decomposition of proteins. The development of the system paved the possibility of development of new high quality biomarkers.
*Biomarker: Molecular information of unique patterns derived from genes and proteins that allow the monitoring of physical changes from genetic or environmental causes.
Professor Lee Kwan Soo’s team (Department of Biological Sciences) composed of Doctorate candidate Han Young Woong, Lee Ho Dong Ph.D. and Professor Park Jong Chul published a dissertation in the April edition of Molecular and Cellular Proteomics. (Dissertation Title: A system for exploring E3-mediated regulatory networks of cellular functions).
Professor Lee’s team compiled all available information of the enzyme that controls protein decomposition (E3 enzyme) and successfully compiled the inter-substrate network by extracting information from 20,000 biology related data base dissertations. The result was the development of the E3Net system that analyzes the related cell function and disease.
Cells have a system that produces, destroys, and recycles proteins in response to the ever changing environmental conditions. Error in these processes leads to disease.
Therefore finding the relationship between E3 enzymes that control the decomposition of proteins and the substrates will allow disease curing and prevention to become much easier. E3 enzyme is responsible for 80% of the protein decomposition and is therefore predicted to be related to various diseases.
However the information on E3 enzyme and inter-substrate behavior are spread out among numerous dissertations and data bases which prevented methodological analysis of the role of the related cells and characteristics of the disease itself.
Professor Lee’s team was successful in creating the E3Net that compiled 2,201 pieces of E3 substrate information, 4,896 pieces of substrate information, and 1,671 pieces of inter-substrate relationship information. This compilation allows for the systematic analysis of cells and diseases.
The newly created network is 10 times larger than the existing network and is the first case where it is possible to accurately find the cell function and related diseases.
It is anticipated that the use of the E3Net will allow the excavation of new biomarkers for the development of personalized drug systems.
The research team applied the E3Net to find tens of new candidate biomarkers related to the major modern diseases like diabetes and cancer.
2012.05.30 View 11620 -
 Creation of Synthetic Antibodies: Professor Hak Seong Kim
Synthetics antibodies which can replace antibodies from humans used as ingredients of medicines have been developed. It can increase the costs to 1/100 of the current costs and is much easier to develop. It is expected that the development period will be shortened from 10 years to 5.
Prof. Hak Seong Kim from the Biology department of KAIST conducted a joint research with Prof. Dong Seob Kim to reconstruct proteins and has succeeded.
The synthetic antibody displays much strength in terms of its productivity, structural formation, and bonding capability, and is thus regarded as an ideal protein. It can replace the antigens that are currently in use. It is expected that Korea will therefore be able to lead the world market for protein medicines which is a 192trillion won industry.
The original antibody has been used for not only treating diseases, but also for various other applications in the fields of medical sciences and biology. However, it is produced through a very complex process involving the incubation of animal cells, and is therefore very expensive. Also, most antibodies are already patented by more developed countries, so a high royalty fee must be paid.
Because of this, many countries including Korea has been concentrating on developing biosimilars copying the antibody medicines for which the patents have already expired. This causes Korea to be behind in the development of antibody protein pharmaceuticals.
Prof. Kim’s research team has focused on the face that the protein existing in some eels are not antibodies but functions as one, and has been successful in developing a synthetic antibody.
The synthetic antibody can be mass produced from the colon bacillus, which allows it to be produced at 1/100 the original cost. It is in a module structure which allows the structuring of the antibody into the desired structure, enabling it to be developed into a protein-based medicine within 5 years.
Together with this, the coherence with the important antigens can be easily controlled, thus allowing for highly effective treatments, less side-effects, high security regarding heat and pH, and the immunogen levels being negligeable. This suggests a very high rate of the antibody being converted into a protein based medication.
The synthetic antibody technology has been tested as a sample for the cure for lung diseases and rheumatism and has been proven to be appropriate. Animal testing will be conducted soon.
Prof Kim said “The original antibodies had a small area allowing the bonding with antibodies, creating barriers for raising bonding strength and structuring. The newly created antibody carries only the strengths and will become a new protein based medicine purely created by Korean technology to replace the antibodies currently used in medications.”
Furthermore, he added that, “The synthesized antibody structuring and designing technology will be widely used in the areas of detecting, diagnosing, and analyzing diseases.”
At the same time, this research result has been published in the Feb 10th issue of the PNAS, and has been supported by the future promising pioneer business program held by the Ministry of Education and Technology.
2012.04.04 View 10122
Creation of Synthetic Antibodies: Professor Hak Seong Kim
Synthetics antibodies which can replace antibodies from humans used as ingredients of medicines have been developed. It can increase the costs to 1/100 of the current costs and is much easier to develop. It is expected that the development period will be shortened from 10 years to 5.
Prof. Hak Seong Kim from the Biology department of KAIST conducted a joint research with Prof. Dong Seob Kim to reconstruct proteins and has succeeded.
The synthetic antibody displays much strength in terms of its productivity, structural formation, and bonding capability, and is thus regarded as an ideal protein. It can replace the antigens that are currently in use. It is expected that Korea will therefore be able to lead the world market for protein medicines which is a 192trillion won industry.
The original antibody has been used for not only treating diseases, but also for various other applications in the fields of medical sciences and biology. However, it is produced through a very complex process involving the incubation of animal cells, and is therefore very expensive. Also, most antibodies are already patented by more developed countries, so a high royalty fee must be paid.
Because of this, many countries including Korea has been concentrating on developing biosimilars copying the antibody medicines for which the patents have already expired. This causes Korea to be behind in the development of antibody protein pharmaceuticals.
Prof. Kim’s research team has focused on the face that the protein existing in some eels are not antibodies but functions as one, and has been successful in developing a synthetic antibody.
The synthetic antibody can be mass produced from the colon bacillus, which allows it to be produced at 1/100 the original cost. It is in a module structure which allows the structuring of the antibody into the desired structure, enabling it to be developed into a protein-based medicine within 5 years.
Together with this, the coherence with the important antigens can be easily controlled, thus allowing for highly effective treatments, less side-effects, high security regarding heat and pH, and the immunogen levels being negligeable. This suggests a very high rate of the antibody being converted into a protein based medication.
The synthetic antibody technology has been tested as a sample for the cure for lung diseases and rheumatism and has been proven to be appropriate. Animal testing will be conducted soon.
Prof Kim said “The original antibodies had a small area allowing the bonding with antibodies, creating barriers for raising bonding strength and structuring. The newly created antibody carries only the strengths and will become a new protein based medicine purely created by Korean technology to replace the antibodies currently used in medications.”
Furthermore, he added that, “The synthesized antibody structuring and designing technology will be widely used in the areas of detecting, diagnosing, and analyzing diseases.”
At the same time, this research result has been published in the Feb 10th issue of the PNAS, and has been supported by the future promising pioneer business program held by the Ministry of Education and Technology.
2012.04.04 View 10122 -
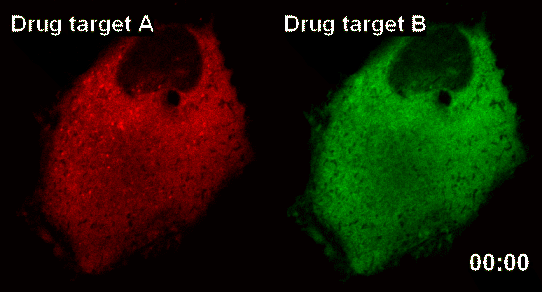 New Technology Developed for Analysis of New Drugs by Using Smart Nano-Sensors
Doctor Sang-Kyu Lee
Doctor Sang-Kyu Lee of the Department of Biological Sciences, KAIST, has developed the technology that allows biological nano particles to be implanted into human cells for monitoring the effect of new drugs in real time from within the cell. It is expected that this technology will boost the ability to weigh the effects and properties of a new drug more quickly and accurately.
Conventionally, the candidate drug was injected into the human body, and then its cells are extracted to analyze the effects of the drugs. The problem with this method was that the cells were analyzed at a ‘dead’ state which made it incredibly difficult to find candidate substances due to uncontrollable side effects. This made the development of new drugs very difficult despite the large costs and efforts invested into its development.
The research team latched onto the idea that nanoparticles can connect to form a large complex. The complex acts as a nanosensor which allows for real time observation of drug target and the drug itself binding.
The team named the nanosensor technology ‘InCell SMART-i’ and was named ‘Hot Paper’ of the September edition of ‘Angewandte Chemie International Edition’ magazine, a world famous Chemistry Magazine.When a new drug injected into the human body, the drug and drug targets are gradually combined, and the smart nanosensor detects in real time the effect of the new drug as shown in the pictures above (shaded spot).
2011.09.19 View 8957
New Technology Developed for Analysis of New Drugs by Using Smart Nano-Sensors
Doctor Sang-Kyu Lee
Doctor Sang-Kyu Lee of the Department of Biological Sciences, KAIST, has developed the technology that allows biological nano particles to be implanted into human cells for monitoring the effect of new drugs in real time from within the cell. It is expected that this technology will boost the ability to weigh the effects and properties of a new drug more quickly and accurately.
Conventionally, the candidate drug was injected into the human body, and then its cells are extracted to analyze the effects of the drugs. The problem with this method was that the cells were analyzed at a ‘dead’ state which made it incredibly difficult to find candidate substances due to uncontrollable side effects. This made the development of new drugs very difficult despite the large costs and efforts invested into its development.
The research team latched onto the idea that nanoparticles can connect to form a large complex. The complex acts as a nanosensor which allows for real time observation of drug target and the drug itself binding.
The team named the nanosensor technology ‘InCell SMART-i’ and was named ‘Hot Paper’ of the September edition of ‘Angewandte Chemie International Edition’ magazine, a world famous Chemistry Magazine.When a new drug injected into the human body, the drug and drug targets are gradually combined, and the smart nanosensor detects in real time the effect of the new drug as shown in the pictures above (shaded spot).
2011.09.19 View 8957