research
Researchers develop a versatile and powerful tool for studying the spatiotemporal dynamics of secretory proteins, a valuable class of biomarkers and therapeutic targets
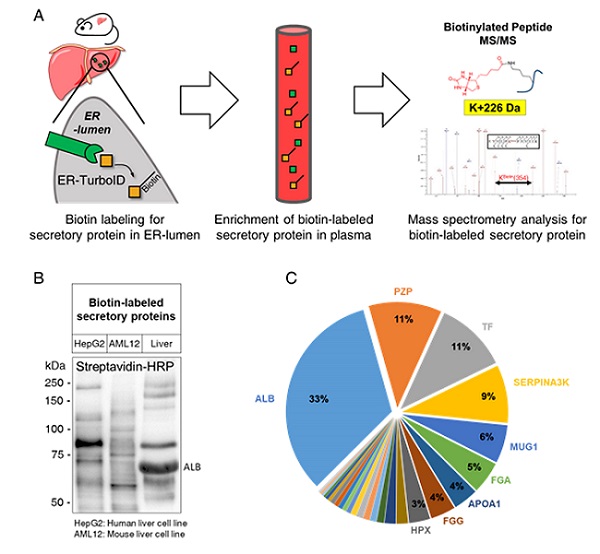
< Figure 1. (A) Schematic illustration for the tracking and identification of tissue-specific secretory protein in live mice (B) Biotin-labeled secretory protein profiles in supernatants of hepatocyte cell lines, HepG2 and AML12, and mouse plasma (C) Composition of liver-derived secretory proteins in mouse plasma >
Researchers have presented a method for profiling tissue-specific secretory proteins in live mice. This method is expected to be applicable to various tissues or disease models for investigating biomarkers or therapeutic targets involved in disease progression. This research was reported in Nature Communications on September 1.
Secretory proteins released into the blood play essential roles in physiological systems. They are core mediators of interorgan communication, while serving as biomarkers and therapeutic targets.
Previous studies have analyzed conditioned media from culture models to identify cell type-specific secretory proteins, but these models often fail to fully recapitulate the intricacies of multi-organ systems and thus do not sufficiently reflect biological realities.
These limitations provided compelling motivation for the research team led by Jae Myoung Suh and his collaborators to develop techniques that could identify and resolve characteristics of tissue-specific secretory proteins along time and space dimensions.
For addressing this gap in the current methodology, the research team utilized proximity-labeling enzymes such as TurboID to label secretory proteins in endoplasmic reticulum lumen using biotin. Thereafter, the biotin-labeled secretory proteins were readily enriched through streptavidin affinity purification and could be identified through mass spectrometry.
To demonstrate its functionality in live mice, research team delivered TurboID to mouse livers via an adenovirus. After administering the biotin, only liver-derived secretory proteins were successfully detected in the plasma of the mice. Interestingly, the pattern of biotin-labeled proteins secreted from the liver was clearly distinctive from those of hepatocyte cell lines.
First author Kwang-eun Kim from the Graduate School of Medical Science and Engineering explained, “The proteins secreted by the liver were significantly different from the results of cell culture models. This data shows the limitations of cell culture models for secretory protein study, and this technique can overcome those limitations. It can be further used to discover biomarkers and therapeutic targets that can more fully reflect the physiological state.”
This work research was supported by the National Research Foundation of Korea, the KAIST Key Research Institutes Project (Interdisciplinary Research Group), and the Institute for Basic Science in Korea.
-Publication
Kwang-eun Kim, Isaac Park et al., “Dynamic tracking and identification of tissue-specific secretory proteins in the circulation of live mice,” Nature Communications on Sept.1,
2021(https://doi.org/10.1038/s41467-021-25546-y)
-Profile
Professor Jae Myoung Suh
Integrated Lab of Metabolism, Obesity and Diabetes Research
https://imodkaist.wixsite.com/home
Graduate School of Medical Science and Engineering
College of Life Science and Bioengineering
KAIST
- No Data